1 Introduction
While it could be said that genetical study of plant development started, in a way, 150 years ago with the publication of Gregor Mendel's paper on ratios in pea genetics, as certain of the phenotypes studied would now be considered developmental alterations, such as fasciation, constricted seed pods, or axial versus terminal pods, the fusion of plant genetics and developmental biology was only to happen long after the rediscovery of Mendel's paper [1–5].
2 Plant genetics and development as separate endeavors
There is no indication that Mendel thought of his characters as developmental alterations, or that he considered his work related to developmental biology as it existed in his day (for example in the work of Payer [6], Fig. 1A, on flower development). The detailed study of plant development began even earlier, not long after the foundation of the Académie des Sciences, with Caspar Friedrich Wolff's 1759 thesis (see Wolff, 1774 [7]), where meristems first were described, with Nehemiah Grew's The Anatomy of Vegetables Begun in 1672 [8] that treated buds as growing shoots (p. 9), or perhaps with Marcello Malpighi's description of a plant embryo in 1679 [9]. The subjects of genetics and development appear to have been separated in the thinking of plant scientists for a very long time afterward, as Bateson points out in his 1894 Materials for the Study of Variation [10], “It has been the custom … to speak of ‘Heredity’ and ‘Variation’ as two antagonistic principles; sometimes they are even spoken of as opposing ‘forces”’ [p. 75]. Bateson agrees with this custom: “In the first examination of the facts of Variation, I believe it is best to attempt no particular consideration of the working of Heredity” [p. 76].
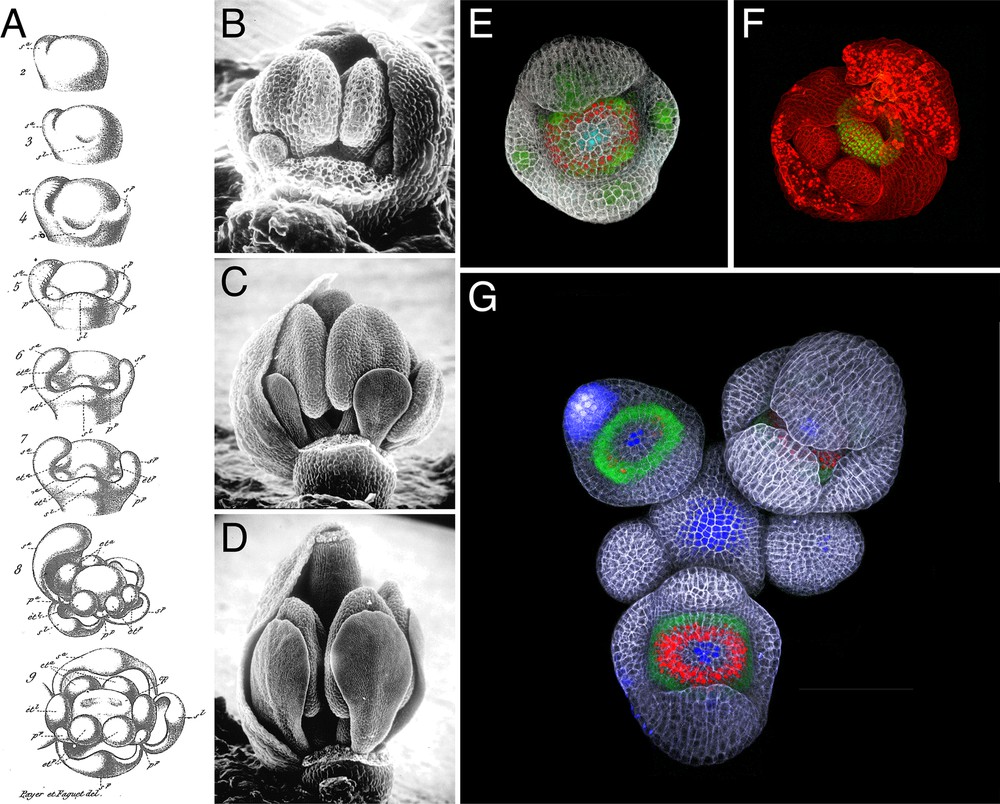
Observation of flower development in Brassicacae, from drawing to live confocal imaging. A. Early flower development in Cheiranthus cheiri as drawn by Payer [6]; drawings show stages equivalent to stages 2–7 in Arabidopsis (stages as described in [71]). B–D. Scanning electron microscope images of developing Arabidopsis flower buds, stages 8 (B), 10 (C) and 12 (D); images courtesy of Smyth and Bowman; images in panels B and D were originally published in [71]. E. Maximum intensity projection of a confocal z-stack of a live stage 4 Arabidopsis flower expressing fluorescent reporters for DORNROSCHEN-LIKE (green; [70]), SUPERMAN (red) and CLAVATA3 (cyan; [72]); cell walls were stained with propidium iodide (grey). F. Maximum intensity projection of a confocal z-stack of a live stage 7 Arabidopsis flower expressing a fluorescent reporter for CRABS CLAW (green); cell walls were stained with propidium iodide (red). G. Maximum intensity projection of a confocal z-stack of a live Arabidopsis inflorescence expressing fluorescent reporters for APETALA3 (green), SUPERMAN (red) and CLAVATA3 (blue); cell walls were stained with propidium iodide (grey).
That this principle was followed by his successors is indicated by the contents of widely used textbooks, such as Steeves and Sussex's Patterns in Plant Development (1972) [11], where genes and mutants do not seem to be mentioned. Mutants are mentioned in the 1989 update of the book [12], and represent the source of much of the developmental information by the 2003 text Mechanisms of Plant Development by Leyser and Day [13]. Thus, there was a transition from considering plant genetics and plant development as unrelated subjects, to considering genetic approaches to be the key to understanding the mechanisms of development that occurred over the past 50 years (see [14]). The use of genetics to understand plant development took far longer, then, than the rediscovery of Mendel's work in the early years of the past century.
To review the history of genetical analysis of plant development, we will take examples from the study of the development of shoots and flowers, a persistent and active subject throughout the history of botanical science. There are earlier examples of the use of developmental phenotypes of plants, such as fasciated plants or double flowers, to study modes of inheritance (for example, White [15,16]; Miyake and Imai [17]; for additional examples in flower development see [18]), and a literature in which developmental mutants are considered as evidence for evolutionary scenarios (e.g., Saunders [19]) – that is, as atavisms. That this is an illogical way to infer evolutionary pathways has been pointed out, in detail, since at least 1900 (Goebel [20]; Leavitt [21]; Arber [22]). In the many works on what we now would call developmental mutants in plants that were published in the 19th and early 20th centuries, and summarized in the compilations of Moquin-Tandon [23], Masters [24], Penzig [25] and Worsdell [26], there was no consideration of whether the phenotypes were inherited (see [18]). The same can be said for Goethe's model for flower development based on abnormal flowers [27] – while this may be the first mechanistic consideration of development, heredity (as expected from the date) plays no role.
3 Early connections
There are nonetheless a few publications in which the potential importance of inherited abnormalities for an understanding of development is highlighted, though without achieving such understanding. For example Leavitt [21] wrote in a review of homeotic variants in plants “The idea of homoeosis unites for descriptive purposes a great number of facts of ontogenesis which, even though they may not at the present juncture point a way to the correct mechanical explanation of development, possess in this connection a considerable prospective value”.
One early, even prescient, use of floral homeotic mutants to propose a model for development of flowers [28] led to the proposal that special hormones were directed to different regions of the developing flower, with the mutations studied leading floral organs to be formed in abnormal locations, thereby missing the hormonal influence. While the direct role of genes in the process is not commented upon, this does introduce a regulatory role for the hormones, if not the genes, whose role is in positioning of organ primordia – the paper is an early example of the use of mutations to develop a mechanistic developmental model.
Another, later example of the use of genetics to understand the mechanism of development in a plant is Stebbins’ work on hooded and awned barley. The mutant form, hooded, has an extra inverted flower that develops on the lemma (one of the bract-like structures that encases a grass floral bud). As Stebbins and Yagil stated [29], “Until recently, the use of genes having pronounced effects on morphology as an aid to the solution of problems of morphogenesis has not been given the attention which it deserves”. They attribute this to the recency of the recognition that genes provide information for the structure of a peptide chain, and on this basis conclude that the goal of plant developmental genetics is to establish the “complete chain of events from primary gene activity to … morphological differences”. To begin this, Stebbins and Yagil made a careful histological analysis of the development of hooded (mutant) and awned (wild type) to find the earliest differences, and found them in more rapid cell divisions in the lemma primordium. The connection between this and the development of a lemma of different structure in the two genotypes was not resolved; in subsequent experiments the connection was sought in changes of peroxidase enzyme activity [30].
4 Regulatory genes connect genetics and development
One interesting aspect of the paper is that there was no consideration that the mutant gene could have encoded a regulatory protein, rather than as a template for an enzyme, and it is likely the idea of regulatory genes ([31]; for plants see [32]), and of positional information (for plants, see [33] where a conceptually more modern view of hooded barley is presented) rather than any technical advance, that permitted the productive study of development using the methods of genetics. Nonetheless, there was some appreciation that the relation of genes to development involved something more than specification of enzyme activities by 1960, for example in Sinnott's book Plant Morphogenesis [34], where he points out that, while little light has been shed on how “genetic factors affect the size, shape and structure of plants,” “Here something more than a series of chemical steps seems to be concerned. Growth relationships are being controlled, and at present we must admit that very little is known about how such control is exercised”.
By 1988 it was clear, at least to one maize geneticist, that the study of mutations might lead to mechanistic understanding of plant development, and that the genes involved would be regulatory: “…it seems reasonable to expect that hierarchies of gene interaction exist wherein certain groups of genes are coordinately regulated to accomplish a particular developmental program… The ultimate choice of developmental program for a cell or a meristem may be under the control of a master switch or regulator genes that regulate entire developmental programs.” [35].
It was conceptual and not technical advances that led to productive use of genetics in plant development. This is manifest in the ABC model of flower development, a positional model of regulatory gene action in the specification of floral organ identity [36–39] that was based like Leavitt's and Brieger's work on homeotic mutants. The origin of this model involved no experiment that could not have been done 50 or even 100 years earlier [40]. It is worth mentioning that the Hooded mutation of barley is now understood at the molecular level – the mutant phenotype was shown by Muller et al. [41] to be due to ectopic expression in the primordium of the extra floret of a transcription factor gene (of the homeodomain family). Even before this, by 1992 and without additional experiments, Stebbins considered flower development to be a matter of successive activations of regulatory factors – a modern view of developmental mechanics [42] that shows in a single author the transition from older to present-day thinking about the relation of genes and development.
Thus, there was a transition from plant genetics and plant development as entirely separate topics, to genetical studies of plant developmental mutants for the purpose of understanding genetic segregation patterns, then study of developmental mutants that did not, because of conceptual failures reveal mechanisms of plant development. We now have in present-day studies a conceptual basis in both genetic and development that have revealed some of the basic processes and principles of plant growth, as well as the detailed mechanisms for many aspects of plant differentiation. The last of these transitions took place between approximately 1960 and 1990. The result is our current, incomplete but powerful understanding of developmental mechanisms in plants and their basis in genetic information.
The later development of the ability for molecular cloning of genes, a technical advance, was the other – but later – half of the equation, as it enabled the nature of genes with developmental phenotypes to be established beyond doubt, and their expression patterns to be studied. In plants, this sort of experiment was considerably aided by another conceptual advance that was as much social as scientific, the development of plant model systems whose intrinsic properties allowed for genetic experimentation, and for facile gene cloning. The ABC model depended on two such systems – Arabidopsis thaliana, whose small genome size allowed map-based cloning (Fig. 1B–D), and Antirrhinum majus, with a highly developed system of transposable element mutagenesis and cloning [39].
5 Flower development in the genetic era
Where has this modern synthesis taken us? An example is in the study of organ specification in flower development, the subject of Goethe's [27], Payer's [6], Leavitt's [21], Brieger's [28] and Stebbins and Yagil's [29] work. The combination of mutant analysis [36–39,43–45], the phenotypes of double mutants (for which there seems to be no precedent – while many double mutants had been studied for flower phenotypes, they seem not before this time to have both been developmental mutants – e.g., Stubbe [46]) and the concept of regulatory genes led by 1991 to the ABC model [37–39]. From the phenotypic analysis of homeotic mutants of Arabidopsis and of Antirrhinum, three classes were found – the A class, which exhibits defects in the outer two whorls of floral organs; the B class, with homeotic transformation of whorl 2 petals into sepals and whorl 3 stamens into carpels; and the C class, in which whorl 3 stamens develop as petals and whorl 4 carpels are replaced by a new, nested flower bud. The ABC model has two major tenets. First, the identity of floral organs is determined by the combinatorial action of these three classes of genes: A class alone specifies sepals, A and B classes combined specify petals, B and C classes combined specify stamens, and C class alone specifies carpels. Second, the A and C class genes are mutually exclusive, and repress each other's expression: in the absence of A class, C class genes are expressed throughout the flower, and vice versa. The ABC genes (APETALA2 [AP2] and perhaps AP1 for A class, AP3 and PISTILLATA [PI] for B class and AGAMOUS [AG] for C class in Arabidopsis) were cloned in the 1990s [47–51], and the ABC model developed into a molecular model. The ABC genes all encode transcription factors of the MADS box family, except for A class gene AP2, which belongs to the AP2/ERF family. Molecular studies confirmed most of the assumptions of the ABC model. B class genes AP3 and PI, for instance, are expressed in whorls 2 and 3, while C class gene AG is expressed in whorl 3 and 4 [47,48,51]. Moreover, ectopic expression experiments corroborated the antagonism between A and C function: plants ectopically expressing AG resemble strong ap2 mutants [52], and plants ectopically expressing AP2 have reduced AG expression in the inner floral whorls [53,54]. Other assumptions of the ABC model proved inexact, however. This is particularly the case with the A class. AP1, for instance, has leaf-like organs, not carpels, in whorl 1 [55], and does not repress AG. Furthermore, while AP2 represses AG, AP2 and AG expression partially overlaps in whorl 3 [56], showing that the second tenet of the ABC model that A and C classes are mutually exclusive is an oversimplification. Even more surprising was the finding that AP2, which promotes petal formation in whorl 2, simultaneously prevents petal formation in whorl 1 by repressing AP3 [57]. A “revised” ABC model taking these newer discoveries into account is presented in [58].
In the early 2000s, the characterization of the SEPALLATA (SEP) genes as a novel, E class of MADS box genes required for the proper specification of the identity of all floral organs [59,60], as did triple mutants of A, B and C class genes [37], confirmed Goethe's hypothesis that floral organs are modified leaves: the combined loss of function of the four SEP genes causes floral organs in each whorl to be replaced by leaf-like structures [59], while the combined ectopic expression of A, B and E genes or B, C and E genes is sufficient to turn leaves into petaloid or staminoid organs, respectively [61,62]. This expansion of the ABC model to include an E class (which also could be considered flower meristem identity, as well as organ identity genes) also led to the proposal of the Quartet model, which postulates that floral organ identity is specified by the action of tetramers of MADS box transcription factors (thus setting AP2 apart from the other ABCE genes): A + E for petals, A + B + E for petals, B + C + E for stamens and C + E for carpels [63]. Despite a large body of evidence that ABCE proteins are able to bind DNA as dimers (e.g., [64–66]), their ability to interact in vivo and form such tetramers was only demonstrated recently [67]. ABCE proteins also interact with a number of chromatin remodeling proteins as well as co-activators and co-repressors, which may contribute to the functional specificity of the quartets [67]. The current working model for the activity of floral organ identity complexes is that MADS box transcription bind DNA as dimers, and dimers interact together to form tetramers, causing DNA looping. The tetramers then recruit additional co-factors and chromatin remodelers to activate or repress the activity of their target genes [68].
The last decade has seen the partial deciphering of the genetic networks downstream of the ABCE genes (reviewed in [58,68], Fig. 1E–F). In sharp contrast with other developmentally important transcription factors that control restricted regulatory networks, ABCE factors appear to have a few hundred to a few thousand targets each. While it is difficult to get a clear picture from so many target genes, some clear trends have emerged from these studies. First, as could be expected for master regulators, a majority of the targets of the ABCE genes are other transcription regulators and genes involved in the metabolism and response to phytohormones. ABCE genes also regulate numerous genes associated with cell proliferation and growth. Second, ABCE genes regulate different targets and promote different developmental programs at different stages of flower development. Third, different ABCE genes share common targets, which is consistent with the fact that the ABCE proteins form heterotetramers to regulate gene expression. Finally, it appears that ABCE genes function not only by promoting flower-specific developmental programs, but also by downregulating genes involved in leaf development, which further confirms Goethe's hypothesis that floral organs are derived from leaves.
Organ identity and its radial pattern is only one (evolutionarily highly conserved) pattern in flowers. Much progress has also been made with radial symmetry of flowers, with floral size regulation, and with the later stages of organ development, leading in total to a detailed but partial understanding of the gene regulatory networks and their downstream genes that create plant form throughout the plant life cycle.
6 Summing up
To a considerable degree, then, Stebbins’ and Yagil's 1966 goal [29] for the field of plant development, that we find the “complete chain of events from primary gene activity to … morphological differences” is well-advanced, and the methods exist to continue to the end. All that was necessary was a combination of technical advances in gene analysis and in microscopy to allow detailed and live imaging of development, gene expression, and protein location (Fig. 1E–G), and just as much, conceptual advances, not least Mendel's recognition and demonstration of the particulate nature of genetic inheritance, coupled with the much later recognition that genes code not only enzymes, but also proteins with regulatory functions.
7 Conclusion: the future
The French Academy of Sciences has a history of 350 years, and its members included Moquin-Tandon, Payer, de Vries, and Tschermak. One hundred and fifty years ago, the basis of inheritance was discovered with experiments using plants, and only 55 years ago were the fundamental concepts of gene regulation put forward. The result already has been progress in the understanding of development, both plant and animal, that in the past 25 years has entirely changed our understanding. What will the coming years bring?
Current methods of genomics and network analysis will allow a much more complete assessment of the gene regulatory networks that underlie plant development in the near future, though there still are technical, and possibly conceptual, barriers to a complete understanding. Among the technical barriers are the daunting prospect of understanding the transcriptional changes in each cell, not each organ, of developing plants, as each organ and the meristems that create them consists of numerous different types of cells, and interacting both through chemical and physical signals. The chemical signaling paths include hundreds of peptides, and hundreds of transmembrane receptor kinases that are still of unknown function, and these must be understood before the mechanisms of plant development can be considered known. A more distant but critical prospect is to understand the connection between gene expression and protein action, and cell biology – how does a particular pattern of gene expression, which might be considered a definition of cell type, lead to the different composition, structure, and activity of cells? A central part of this understanding will come from comprehension of protein interactions, and from the emergent properties of protein machines that create cellular activities, and interact with non-protein cellular structures, such as membranes. Genetics has played a large role in this area to date, but advances in structural and synthetic biology will be needed to complete this program. And after this, the interactions between cells that dynamically regulate cellular activities will also have to be catalogued and developed into causal networks. The large number of gene interactions, protein interactions, and interactions between cells involved make it abundantly clear that development of biological methods and concepts must be accompanied by development of computational methods to encompass and test the causal interactions between the myriad of parts within a cell, the myriad cells, and the interactions of the plant with its environment. The development of a set of computational approaches, without which we will never be able to claim understanding of plant (or any other) development, is perhaps the next great challenge – such that our understanding of regulatory genes, that is to say the extraction of the informational content of organisms, can be matched by its cognate process, the construction (in silico first, then in vivo) of organisms from their informational content. This will be a necessary proof of principle, and also a type of completion of the research program started by Mendel – the deconstruction, then reconstruction, of life based on the principles of genetics whose first elucidation began 150 years ago.
In the field of plant development, however, one thing has never changed, and is unlikely to change: as stated by the plant physiologist Karl von Goebel, “For an experiment one needs a plant, a flower-pot with earth, and a question” [69].
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
The flower development work in the Meyerowitz laboratory is funded by the U.S. National Institute of General Medical Science (R01 GM104244), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation (GBMF3406).


