1 Introduction
For a long time, the genome was considered as a relatively compact unit that did not undergo any major changes in the course of many generations. Over the past few decades this view has been considerably modified due to an increasing understanding of the structure of genomes and the discovery of their plasticity.
The evolution of eukaryotic genomes takes place via rearrangements that can sometimes occur on a large scale. We might therefore speak of macro-rearrangements or, on a smaller scale, micro-rearrangements. These rearrangements can result from a gain or loss of DNA, but can also be simple reorganizations of the genome.
Among the macro-rearrangements are polyploidisations (duplications of the totality of the genome) and segmental duplications. Following a polyploidisation, different rehandlings can occur; some of the duplicated genome is lost, while the conserved part can undergo multiple rearrangements (see the article by Jaillon et al. in the present issue, [1]). Segmental duplications, on the other hand, are produced at a frequency that is probably higher than hitherto imagined. They can be large in size; what happens to them depends on selective forces and drift (see the article by Koszul and Fisher in the present issue, [2]).
To understand micro-rearrangements, we must look at the content of a genome. Three kinds of sequences are found in eukaryote genomes: highly repeated sequences, middle repeated sequences and unique sequences. Highly repetitive sequences include satellite sequences (heterochromatic), minisatellites and microsatellites (occurring throughout the genome); middle repetitive sequences include transposable elements and certain multigenic families, such as histone genes; finally, unique sequences correspond to genes. The proportion of the different types of sequences varies from one organism to another. Moreover, each of these sequences has a certain plasticity, giving rise to plasticity in the entire genome.
The number of repetitions of “satellite” sequences (satellites, microsatellites and minisatellites) rises or falls due either to slippage of the polymerase during replication or to unequal recombination. These sequences can have an important role in the plasticity of genomes; they may play a part in the origin of segmental duplications (see the article by Koszul and Fisher in the present issue, [2]), but also in the evolution of genes, as is the case for neurodegenerative diseases in human beings, also known as triplet diseases.
For middle repetitive sequences, only the transposable elements will be presented here. For multigenic families, an example of evolution is given in the article in the present issue by Wajcman et al. on the globin super-family [3]. Transposable elements may be defined as DNA sequences that can (or have been able to) move and/or multiply within a genome. They represent a variable part of genomes: approximately 3% of the genome in Saccharomyces cerevisiae yeast, 14% in Arabidopsis thaliana, 20% in drosophila (Drosophila melanogaster) and 44% in man (Homo sapiens). They are highly diverse, but we can define four large groups: the transposons that transpose via a DNA intermediary; the retroposons that transpose via an RNA, and for which integration into the genome is coupled with reverse transcription, and within which we can distinguish Long Interspersed Nuclear Elements (LINE) and Short Interspersed Nuclear Elements (SINE); finally, retrotransposons, which also transpose via an RNA, but where a cDNA is formed. Among the latter, we can distinguish the elements for which the integration of the cDNA takes place due to an integrase and which have at their extremities Long Terminal Repeats (LTR), and those for which the integration takes place due to a tyrosine recombinase, and which can show Split Direct Repeats (SDR) or Inverse Terminal Repeats (ITR) at their extremities.
2 Impact of transposable elements on genomes
2.1 Chromosomal rearrangements
Transposable elements play a large part in the plasticity of eukaryotic genomes. As sequences repeated in the genome, they can be at the origin of chromosomal rearrangements through ectopic recombination, that is, recombination between homologous sequences in nonhomologous chromosomal sites. If recombinations occur between the sequences present in the same chromosome, chromosomal inversions or deletions can then be observed. If they are produced between elements present in different chromosomes, translocations (balanced or unbalanced) can be observed. According to the chromosomal part involved, macro or micro rearrangements may take place. In the drosophila, the Antp73b allele with the dominant effect of the homeotic gene Antennapedia, characterised by the presence of legs instead of antennae, is due to a micro-rearrangement of this kind [4]. It is caused by an ectopic recombination between two elements of the LINE type, leading to a reciprocal exchange of the first exon of the Antennapedia gene with the first exon of the rfd gene (responsible for dominant phenotype), the function of which is not known (Fig. 1).
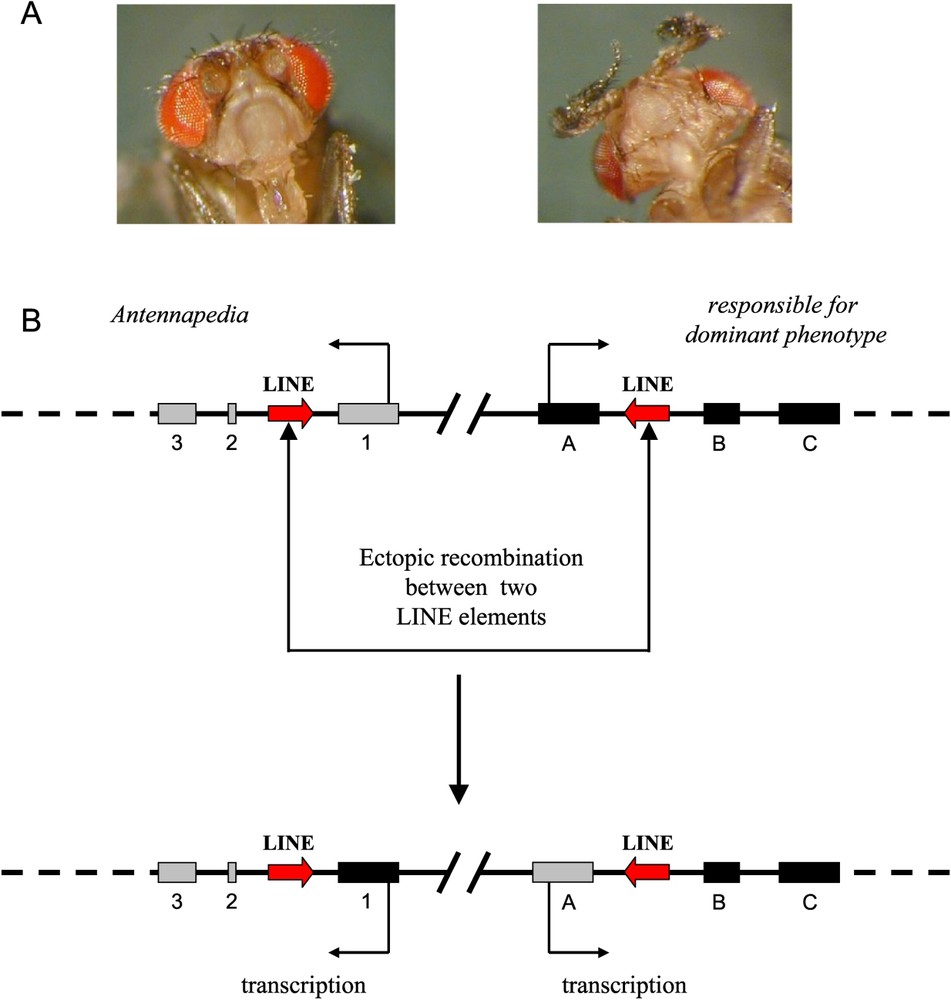
Exchange of exons by ectopic recombination between two elements of the LINE type. A. Left, wild phenotype corresponding to the homozygote genotype Antp + /Antp+; right, Antennapedia phenotype corresponding to the heterozygote Antp + /Antp73b. B. Diagram of the exchange of the first exons between Antennapedia and rfd genes (from Schneuwly et al., 1987 [4]).
2.2 Modification of genes or their expression
Transposable elements can also play a part in the plasticity of genomes due to their mobility. They can be inserted in genic or intergenic regions. The consequence of their insertion into genes, though often harmful, can sometimes be beneficial or neutral. In the latter case, the insertion can be fixed by genetic drift. The systematic study of the human genome carried out since it was completely sequenced has meant the importance of transposable elements in the evolution of the transcripts of genes can be quantified more precisely. A study of almost 14 000 human genes [5] shows that 4% of them contain sequences in their coding region that have similarities with transposable elements. Over 89% of these elements seem to correspond to insertions within introns that have then been recruited as exons, with the rest resulting from insertions in exons.
If the frequency in coding sequences may seem small, it increases if we look at cis-regulating regions of genes. The importance of insertions in cis-regulating regions of plant genes was understood as early as 1994. White et al. [6], with the LTR retrotransposon Hopscotch, and Bureau et al. [7,8], with the transposons Tourist and Stowaway, showed that over 120 plant genes then present in the databases contained a transposable element in their cis-regulating regions. Since then, the sequencing of complete eukaryotic genomes has allowed these phenomena to be quantified more precisely. In human beings, the study of 12 179 loci contained in the RefSeq database (http://www.ncbi.nlm.nih.gov/RefSeq/) reveals that 27% of genes contain a transposable element in their 5′ or 3′ transcribed but not translated region; in mice, the figure is 18% of genes out of 10 064 loci studied [9]. Insertion of a transposable element in 5′ of a gene can create a new promoter that will, in certain cases, acquire a new specificity, or else will allow an alternative promoter to be created. Different cases of this kind have been described in primates and in man. The new promoter often corresponds to a LTR of retrotransposon, since the LTR contains all the sequences needed for transcription to be initiated.
In the cases outlined above, the impact of the transposable elements on the genome could be said to be “indirect”; in fact, more and more examples have been described in which it is the own function of the transposable element, namely its capacity to transpose, that is “recruited” by the genome.
2.3 Recruitment of transposable elements
2.3.1 Elements of the LINE and SINE type
In the drosophila, the telomerase function does not exist. This function is carried out by two transposable elements, the LINE TART and the SINE Het-A. After each replication cycle, the transposition of a Het-A element or a TART element is occurred to a specific site, at the extremity of the chromosomes. In fact, reverse transcription of one of these elements takes place due to the transcriptase reverse of the LINE TART [10]. Similarly, in the silkworm, Bombyx mori, an element of the LINE type is also inserted at a specific site, at the telomere level. However, in this species, the telomerase function still exists. The same situation is found in the green seaweed, Chlorella vulgaris. In fact, the reverse transcriptase of a LINE and telomerase function in a similar way and, structurally, the reverse transcriptase domain of the telomerase shows similarities with reverse transcriptases of retrotransposons [11]. This suggests a similarity between reverse transcriptases of telomerases and retrotransposons, and raises the question of the common ancestor of the two reverse transcriptases.
2.3.2 Elements of the transposon type
Transposons transpose according to a cut-and-paste mechanism: the original copy is excised from the ‘donor’ site and is then inserted elsewhere in the genome at the target site. This takes place thanks to an enzyme, the transposase, which is coded by the complete or autonomous element. The transposase can also act in trans, and mobilise defective elements, on condition they have conserved Inverted Terminal Repeats (ITR), which are found at the extremities of these elements (Fig. 2A). The two following examples show how this capacity to excise can be recruited by the genome.
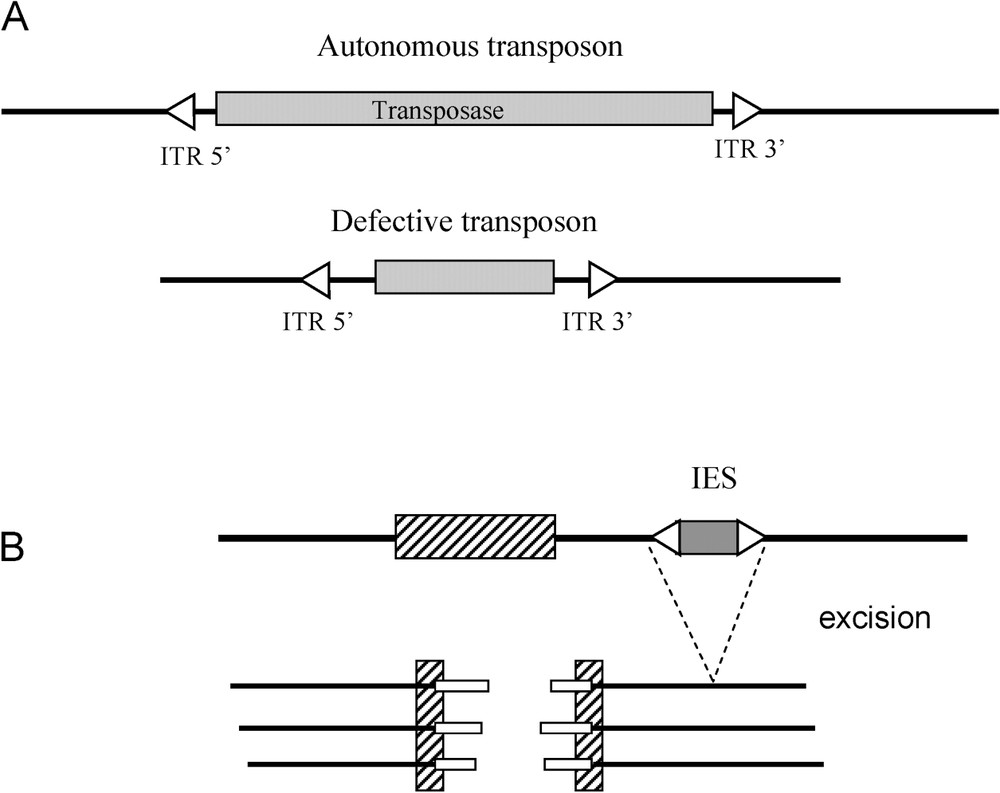
Recruitments of transposable elements. A. Diagram of an autonomous transposon and a defective transposon. ITR: Inverted Terminal Repeat sequence. B. Rearrangements of the macronucleus in ciliates. IES: precisely eliminated internal sequences. Hatched box: imprecisely eliminated genomic sequences. White boxes: new telomeres.
In ciliates, two nuclei are present, the micro-nucleus and the macro-nucleus. The micro-nucleus is diploid and transcriptionnally inactive. Its role is to transmit genetic information during reproduction. The macro-nucleus presents fragmented chromosomes and is transcriptionnally active. The macro-nucleus contains short molecules of linear DNA that derive from chromosomes of the micro-nucleus through fragmentation and elimination of internal sequences (IES sequences). Telomeres are then added to the extremities of these fragments. Last, the macro-nuclear chromosomes are amplified, with a final ploidy of about 1000. The mechanisms at the origin of the excision of the IES are close enough to the mechanisms of the excision of transposons for Seegmiller et al. [12] to suggest that the IES are former transposons that have lost a part of their internal sequence, and have thus become defective (Fig. 2B). In parallel with the loss of these internal sequences, a transposon coding the equivalent of an ‘excisase’ may have become immobile (loss of ITR) and been placed under the control of a promoter of the host (for a review, see [13]).
Another case of recruitment of the enzymatic function is given by the immune system of certain vertebrates. The diversity of immune system response in vertebrates is generated at the lymphocyte level in the course of their development, following the rearrangements of DNA at the level of the genes coding the immunoglobulins and the T cell receptors (Fig. 3). The process leading to these rearrangements has been called V(D)J, Variable (Diversity) Joining. V(D)J necessitates the activity of two enzymes, RAG1 and RAG2. These two enzymes are coded by two genes, rag1 and rag2. RAG1 and RAG2 have the capacity, on the one hand, to recognise specific sequences corresponding to combination signals placed between the V, D and J and, on the other hand, to cleave the DNA at this level, due to: (1) the structure of these recombination signals (similar to ITR); (2) the capacity in vitro of RAG proteins to allow intra- and inter-molecular transposition of integrated DNA sequences between two recombination signals [14]; and (3) the repair mechanisms of double strand breaks in DNA after excision of the fragments V(D) or (D)J that are similar to those observed for the repair of breaks in double strands of DNA after the excision of transposons [15–17]. It seems highly likely that the rag1 and rag2 genes derive from the sequence coding the transposase of a transposon. Later, Kapitonov and Jurka proposed that rag1 gene derived from Transib transposase [18].
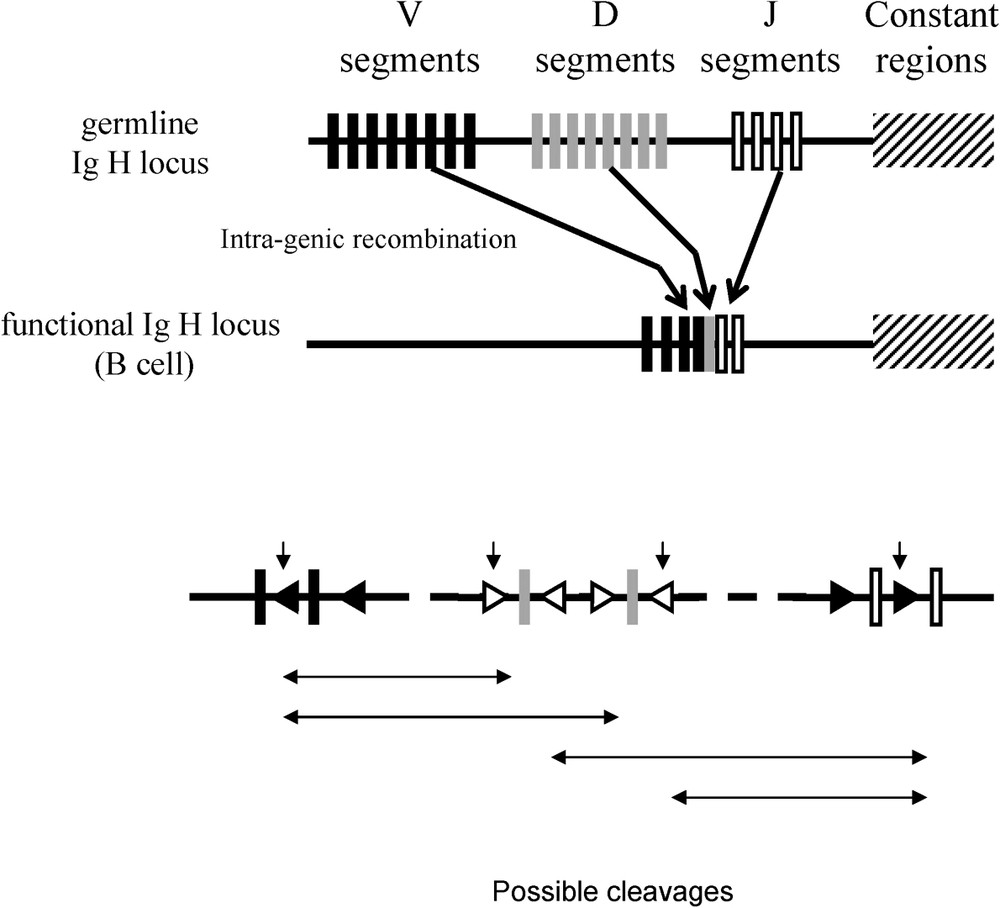
Intragenic rearrangements in the somatic line in order to generate the variability of the range of immune responses (from Sadofsky 2001, [17]).
3 Evolution of genic sequences
We have just seen that, in diverse ways, transposable elements play a major role in the evolution of genes. However, other mechanisms take part in the evolution of genic sequences. This is the case, for example, of pseudogenes, which are very often found in eukaryote genomes. These pseudogenes can be created by the duplication of a fragment of DNA or are issued from the reverse transcription of an RNA. In the latter case, where the cis-regulating and promoter sequences are absent, it is transcriptionnally inactive, except if they acquire new promoter sequences. Transposable elements of the LINE type can play a role in this mechanism of retroposition; in fact, it has been shown, for LINE L1 in particular, that its reverse transcriptase can reverse-transcribe all polyadenilated RNA [19]. The number of pseudogenes can be large, but varies according to the organism; in man and the nematode, it is estimated that about 10% of known genes possess a pseudogene, while in the drosophila, only 1% of genes have them. This variability between organisms is also found in the origins of these pseudogenes; in mammals duplication and retroposition play an equivalent role, while in Saccharomyces cerevisiae yeast they are issued from duplication alone. The evolution of the number of genes (genes in the strict sense or pseudogenes) is not the only level of plasticity within these sequences. The analysis of proteic sequences and their tri-dimensional structure has shown that many proteins were constituted in domains. These proteins, defined as mosaic, are particularly abundant in metazoas. The study of genes that code mosaic proteins shows there is a high correlation between the organisation of the domains and the intron-exon structure. Each domain is coded by one or several exons that limit the domain, suggesting that this type of protein may have been created by exon shuffling. The hypothesis of modularisation of proteins involves three stages: (1) the insertion of introns in a position corresponding to the limit of the domains; (2) duplication of the whole interior of the inserted intron; and (3) transfer towards other genes by ectopic recombination at the intron level (see [20,21]). However, a further mechanism, the retroposition via transposable elements of the LINE type, can play a part in exon shuffling [22,23]. The evolution of genes through exon shuffling means the excision of introns and exon-splicing must be carried out correctly, which shows that this kind of evolution is closely linked to the evolution of introns, and especially of the “spliceosome”.
4 Conclusion
The plasticity of eukaryotic genomes results from all the mechanisms listed above. Moreover, these mechanisms are not mutually exclusive: micro-rearrangements, for example, can occur after macro-rearrangements. Similarly, the different kinds of sequences can interfere with the evolution of the others; this is the case, for instance, of transposable elements and genes. Thus we can see that the eukaryote genome is highly fluid. The fluidity of genomes gives rise to genetic variability, which will be exploited by evolutionary forces (selection and drift). Many genetic innovations are evolutionary failures that can only be observed in the laboratory in model organisms at present, as is the case for the mutation leading to the Antp73b allele of the Antennapedia gene described above. In nature, such a mutant would be eliminated by natural selection. Others can be positively selected or else be neutral and fixed by drift. Selection can be envisaged on two levels: on the individual level (the whole genome), the best adapted individual leaving behind the most descendents (Darwinian selection) or on the level of the DNA sequence (notion of selfish gene). Transposable elements and microsatellite sequences have often been taken as illustrations of Dawkins' theory of the selfish gene [24]. As we have extensively described the impact of transposable elements on the evolution of genomes, can we consider that these sequences are always selfish? Pinsker et al. [25] propose an evolutionary scenario for the development of a transposable element within the genome. The first step consists in its arrival within the new genome, which can take place by horizontal transfer. This element is then amplified through transposition until the regulation mechanisms of transposition are in place. Once the element is immobilised, three scenarios are possible: the element is lost; it is transferred to a new genome for horizontal transfer or it is recruited by the genome. It is in fact during its multiplication phase that the transposable element can be considered as selfish DNA. When it is recruited, however, it is maintained in the genome by the advantage it gives to the genome containing it.
The comparative study of eukaryotic genomes allows us to bring out the mechanisms at the origin of the fluidity of genomes. These mechanisms are not large in number, but together they can produce an infinity of combinations. This fluidity means that genetic variability can be generated that is, so to speak, infinite. Genetic innovations maintained during evolution show, on the one hand, that evolution is not necessarily parsimonic and, on the other hand, that it has no aim.


