1 Introduction
In this paper we construct and analyze a fully continuous individual-based model (IBM) for cancer invasion.There have been many recent studies of tumor growth in spatial settings, including [1–11]. In our model of spatial tumor growth our aim is two-fold: first we construct an IBM founded on stochastic events, which yields readily interpreted graphical visualizations of tumor growth. Second, we transform the IBM to a system of partial differential equations (PDE) for the continuum densities of the IBM elements (in the limit of large numbers of particles). Then we obtain the numerical solution of the PDE, and this solution is qualitatively compared to the IBM output, in order to validate the IBM approach.
Individual-based models in mathematical biology have been used extensively recently, because increased computational capability allows the possibility of emulating in virtual worlds the huge variability of individual behavior. The consideration of different kinds of individuals is natural in IBM technology, with heterogeneity of individuals embedded in object-oriented programming. A frequent assumption in differential equations models of population dynamics is the so-called mean-field description, which assumes that individuals are homogeneous in a spatial context. In such models the influence of environment and the interactions between individuals are averaged throughout space, and spatial dependence is simplified or omitted. For many biological processes, however, spatial heterogeneity of individuals is an essential feature in understanding population behavior, and IBM approaches are advantageous for inclusion of spatial heterogeneity (a basic reference for IBM is [12]).
Although the visual aspect of IBM simulations are appealing to non-experts, IBMs still lack a general theoretical framework to interpret their output, which are different every run. In order to bridge the gap between computer simulation and mathematical description of our tumor invasion IBM, we use a set of scaling techniques to derive large-scale descriptions from the individual behavior. General references for this approach are [13] and [14], and the seminal paper [15].
The biological aims of our study are to develop a mathematical description of tumor growth with emphasis on spatial invasion into surrounding extracellular matrix. The biological complexity of this process invokes multi-scale levels – sub-cellular, cellular, and tissue. Mathematical frameworks provide a medium for the biological understanding of tumor growth through integration of these levels. The larger biological aim of our study is to develop a reliable predictor of tumor growth amenable to computer simulations specific to individual patients.
In this paper we focus on a hybrid discrete-continuous model recently presented by A. Anderson in [16], which we use as starting point. Related developments and numerical treatments can be found in [17]. We refer the reader to these references for the biological considerations and features of the model. We build and study an improved model for the same phenomenon. The improvements to Anderson's model are:
- (1) Our model does not have a rigid discrete lattice spatial structure, but instead provides a more biologically realistic continuum spatial setting for cell growth and proliferation;
- (2) Our model includes a cell age structure connected to a PDE formulation to describe individual cell behavior, and in particular allows inclusion of the phases of the cell cycle correspondent to cell age;
- (3) Our model is convertible to a completely PDE formulation (the model in [16] was only partially convertible) and is advantageous for computational simulations;
- (4) Our model can be analyzed theoretically and the significance of the parameters can be quantified.
As in [16], we analyze conditions under which tumor growth exhibits fingering, and in order to study this phenomena, design simulations with different initial MM distributions: homogeneous and heterogeneous.
With the aim of validating the individual-based approach, we scale-up the IBM (Lagrangian framework) to obtain a density-based Eulerian procedure. We construct a set of PDEs equivalent to a large number of runs of the computer simulations. We obtain numerically the solutions to these PDEs and qualitatively compare the results of both approaches with special interest on fingering patterns. We have made extensive numerical simulations, which strongly indicate that both approaches show very similar results. A formal validation of the equivalence of the two approaches will be explored in future work.
The organization of the paper is as follows: Section 2 contains a brief description of the biological features which are taken into account for the construction of the model; in Sections 3 and 4 we describe the tumor invasion IBM and its equivalent formulation in terms of PDEs. Section 5 focuses on the numerical analysis of the PDE model. Section 6 concludes with a discussion of results and conclusions.
2 Biological background
Tumors develop as mutations occurring in key regulatory genes of cell proliferation. According to [16,18], one of the most important mutations involves the p53 gene, the so-called Guardian of Genome, which is found in mutated form in over half of all cancers. The p53 protein controls three cellular functions: proliferation, death and DNA repair, so that a loss of p53 function due to mutation allows for the propagation of damaged DNA to other cells.
The aim of this paper is to develop a model for the growth of a generic solid tumor for which we will assume a blood supply has been established. We are especially interested in the influence of cell adhesion (i.e., cell–cell and cell–matrix adhesion) in the tumor shape, as this is a key question in the invasive process. Molecules in charge of cell adhesion not only regulate cell migration, but proliferation and differentiation as well.
Following [16], we restrict the model to the behavior of four principal elements: tumor cells (both proliferating and quiescent, and classified by mutation phenotype), extracellular matrix macromolecules (MM), matrix degradative enzymes (MDE) (which break down the MM), and oxygen (which supplies essential nutrients from the surrounding vasculature). Tumor cells proliferate, enter quiescence, or die according to the concentration of nutrients provided by the capillary network, which is simulated by the diffusion of oxygen. Proliferating cells have the ability to migrate, but quiescent cells do not and there is experimental evidence that cells move towards high MM concentrations. Some regions may become more nutrient deficient, until eventually cells can no longer survive, degenerating into necrotic regions. Macromolecules disappear progressively with time as they are degraded by tumor cells, the diminishing rate being proportional to the MDE concentration. MM are non-diffusible since they are bound within the surrounding tissue. Matrix degradative enzymes undergo several processes, and such enzymes facilitate degradation of extracellular matrix allowing adhesion and attachment of tumor cells. They are produced by cells, diffuse, and decay naturally. Finally, oxygen diffuses, is produced by MM, is consumed by cells and decays naturally.
3 Description of the IBM for tumor invasion
In this section we describe the way in which the behavior of the four kinds of particles that we consider in our model have been taken into account to built the IBM. For simplicity we assume the tumor is contained in a 2-dimensional spatial region, although our methods are applicable to the 3-dimensional case.
3.1 Description of the particles
3.1.1 Tumor cells
The tumor consists of populations of proliferating and quiescent cells. Proliferating cells are capable of growing, dividing, entering quiescence and becoming necrotic. We consider the possibility for a cell to mutate only at division, and the probability of mutation is taken to be . We consider four types of mutations, so the number of actual phenotypes is five with the wild-type is also taken into account. This scheme of mutation is a simplified treatment (as in [16] and [17]). Our simplified framework of proliferating mutation classes is readily adapted to a greatly expanded number of phenotypes, but with greatly increased computational cost. Nonetheless, we believe this simplification to be representative of the effects of mutations.
We track progress through the cell cycle with cell age, and newly divided cells start with cell age 0. The scheme of mutation is linear, so a mutation results in a passage to the next mutation phase. The higher the mutation phenotype, the higher the cell's aggressiveness. This aggressiveness is due to the possibility of higher motility, lower proliferation age, higher oxygen uptake and greater MDE production.
Proliferating cells divide through a distribution of ages, and the probability for a cell to divide at a given age depends both on its age and the oxygen concentration nearby. Based on [17] we choose the proliferation rate in the IBM as:
| (1) |
An absence of oxygen can also lead cells to the quiescence state, and the return to the proliferating state is only achieved if sufficient oxygen becomes available again. In the IBM, both changes are modelled with transition rates (from proliferation to quiescence) and (vice versa). As such, the transition rate is assumed to roughly decay with oxygen concentration c, whilst behaves the opposite way. With these ideas in mind, and using the formulas from [17]:
| (2) |
Experiments show that cells have random motility. A Brownian motion seems suitable to reproduce this movement and the uncertainty inherent in the process of modeling, as explained in [19]. In the IBM, at every time step, each cell jumps in a random direction. The jump length is distributed as a Gaussian with mean zero and standard deviation:
Cells are attracted by high MM concentrations, in a phenomenon called haptotaxis in which the intensity of this attraction is assumed to be proportional to the concentration gradient. Haptotaxis is, from the mathematical side, similar to chemotaxis (see [21]) and, in general, equivalent to any motion whose speed is proportional to the gradient of concentration of any substance, particle or individual type. In our model the intensity of the haptotaxis is described by a corresponding coefficient . As before, is the mutation type, and stands for the haptotaxis intensity for the corresponding mutation, assumed to grow with the mutation type. The constant is the intensity of the wild-type haptotaxis. In the IBM, at every time step, each cell jumps a length
| (3) |
3.1.2 Macromolecules (MM)
Macromolecules form the tissue surrounding tumor cells and are crucial in our model, since they induce the haptotaxis phenomenon resulting in tumor fingering. In particular, their initial spatial distribution determines the evolution of the tumor spatial pattern. Following [16,18], we will consider different types of initial MM spatial distributions, homogeneous and heterogeneous as explained below.
MMs undergo degradation produced by the MDE particles, and the intensity of their degradation is proportional to the product of concentrations of MDE and MM. This will produce a so-called reaction term in the equivalent PDE formulation (see [22,23]). To reproduce this phenomenon in the IBM, the probability for a MM particle to disappear at any time step is taken to be proportional to the concentration of MDE nearby. Another way to model this process is by considering the number of collisions per time unit between MDE and MM particles. We believe both ways are equivalent for sufficiently small time steps. More precisely, the probability for an MM particle at position to disappear at any small enough time step dt is , where is a constant related to the intensity of the reaction and is the concentration of MDE. MM particles are assumed bound, and thus have no motion at all, at least in comparison to tumor cells.
3.1.3 Matrix-degradative enzymes (MDE)
MDE have a natural tendency to spread throughout the tumor environment and experimentally exhibit a random motion. As for cells, a Brownian motion is implemented for MDE particles in our IBM. The deployment of this motion is exactly the same as with cells, the intensity coefficient now called . MDE are generated by cells, and the production rate is assumed to be proportional to cell concentration. To properly emulate this in the IBM, at each time step the probability for an MDE particle to be created is proportional to the surrounding cell concentration, more precisely the probability is taken to be
| (4) |
3.1.4 Oxygen
Oxygen spreads throughout the environment, is produced at rates proportional to the MM concentration, is consumed by cell uptake, and decays naturally. All these processes are incorporated in the IBM in the previously described manner. The Brownian motion intensity is called , cell uptake rate is , with a constant, the constant decay rate is and the constant production rate is .
3.2 IBM computational scheme
We model the tissue as a square (we allow the particles to move outside this square, but we will focus on the inner region). Summing up, we model the evolution of 4 types of individuals: cells, MM, MDE and oxygen. We consider 5 phenotypes for cells: wild-type plus mutation types 1, 2, 3 and 4 and consider a linear mutation scheme, so each mutation makes a cell increasingly aggressive, according to the values given in Table 1.
The IBM computational scheme
| Phenotype | Oxygen uptake | MDE production | Brownian coefficient | Haptotaxis coefficient |
| Wild type | ω | κ | D | ρ |
| Mutation 1 | 3ω/2 | 3κ/2 | 3D/2 | 3ρ/2 |
| Mutation 2 | 2ω | 2κ | 2D | 2ρ |
| Mutation 3 | 5ω/2 | 5κ/2 | 5D/2 | 5ρ/2 |
| Mutation 4 | 3ω | 3κ | 3D | 3ρ |
As the cells evolve through mutations, they became increasingly aggressive, due to stronger motility, stronger oxygen uptake and stronger MDE production. We remark that our scheme is space continuous, avoiding limitations of discrete approaches. We think this is important in order to connect the discrete simulation to the PDE approach. See Fig. 1 for a schematic flow chart of the IBM algorithm.
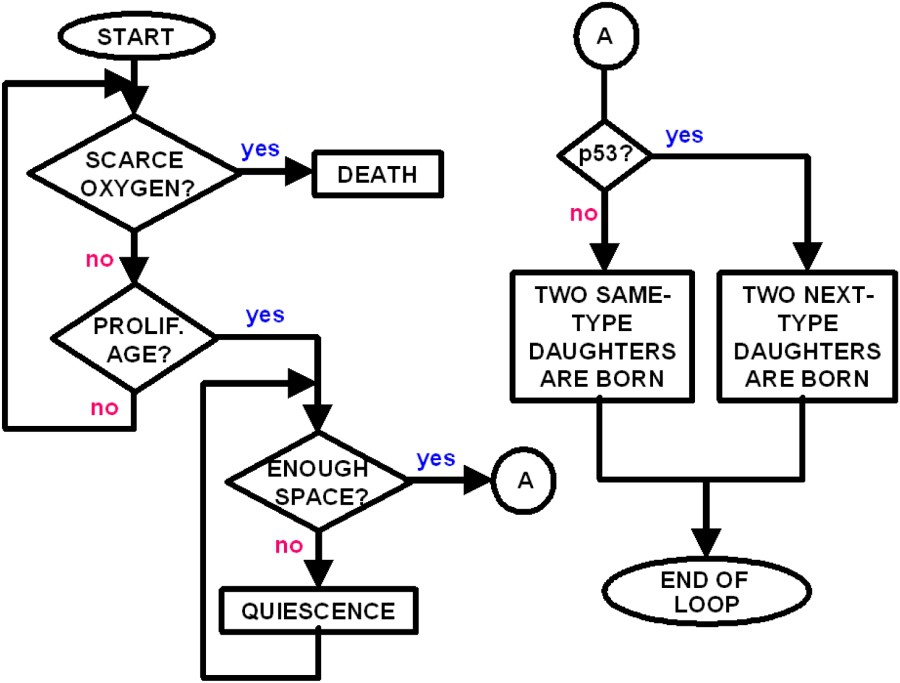
Summarized flowchart of the IBM algorithm.
3.2.1 Initialization stage
We are interested in the development of tumor fingers, which form as a consequence of the attraction cells experience for macromolecules. To discern the influence of initial MM spatial distributions on the development of tumor patterns, we carry out two different experiments, one with MM homogeneously distributed and the other heterogeneously distributed.
Cells.
In both experiments all cells are initially spatially distributed according to the density formula:
In this way, we model a pre-invasive stage, where cells are initially present in the neighbourhood of the domain center, with lower densities along any radius starting at the tumor center. Initially we assume that about 100 cells are present. Also, we assume that all cells are initially proliferating with an age distribution between 0 and 2 days.
Macromolecules.
As we previously indicated, we will carry out two experiments corresponding to homogeneous and heterogeneous MM distributions, respectively:
- (A) Macromolecules homogeneously distributed along the domain at with a fixed density of 0.3;
- (B) Macromolecules intentionally heterogeneously distributed over the same domain at . This heterogeneous distribution has two maximum peaks in the upper-right and the lower-left zones, more exactly at points and . We choose this distribution to intensify the expected consequences of haptotaxis. The density is
Matrix-degradative enzymes.
The density of MDE is taken to be half the proliferating cells density in both experiments.
Oxygen.
The density of oxygen is taken to be 10 times the MM density in both experiments.
Our parameters are chosen for mathematical illustration as well as for biological realism (as in [17]). Biological validation of these choices requires the experimental support of current efforts [24]. Simulations were run from through . In order to be reasonably sure that the qualitative results of our simulations were representative, we run the simulation 200 times. The results of these simulations were quite similar.
3.3 Routine to compute local densities and gradients
The local densities and gradients of oxygen, cells, MDE and MM are computed in several subroutines of the IBM code. In order to measure these quantities, we assume that particles have a limited perception of the surrounding neighbourhood, so they can measure, at least roughly, the local densities and gradients. We assume this perception to be limited to a rectangle whose axis lengths are units in x and y, respectively. This concept of limited perception is quite usual for IBMs in mathematical biology, [25], due to their description of local interactions.
For each particle k, which may be a cell, MM, MDE or an oxygen particle, let be its position in the 2D space. For example, let denote the number of MM particles, perceived by individual k on its right side, i.e., inside the rectangle
Several values of and were tested, but in the end we found the value to be reasonable, because this value is large enough to include a high number of particles inside, and low enough to be small as compared to the whole square. The choice is a trade-off between both circumstances. The high number of particles considered slows down the computation loop, because the previous measure of gradients and concentrations must consider each particle individually. The execution of our simulations took about 24 hours on a Pentium III processor. The results of the simulations can be seen in the figures. The interested reader can also check some videos (.avi format) of the simulations on http://dmaii.etsii.upm.es/~cells.
4 Continuous PDE formulation for the IBM
In order to describe the algorithm underlying the IBM in mathematical language, we will adapt the stochastic approach described in [26] and [15]: the evolution of each particle can be written as a stochastic differential equation for the motion together with Poisson processes for the birth and death. Then, under some assumptions, a passage to the limit when the number of particles tends to infinity leads to a PDE for the density of each kind of particles. We also refer to [14] and [27].
4.1 General description of the procedure
When dealing mathematically with the IBM, we must distinguish two kind of processes: motion and demography. Motion processes are only related to the spatial evolution of IBM particles, while demographic processes include births, deaths and transitions between different types of particles. Roughly speaking, motion processes will include transport and Brownian phenomena and will be described by stochastic differential equations (SDE), while demographic phenomena will be described by Poisson processes with certain intensity rates key to the model. For a detailed explanation on SDEs and Poisson processes we refer the reader to [28] and [29].
We describe the general procedure in two steps:
Step 1. First of all we consider a general IBM where only two-dimensional motion takes place and only one kind of particle appears. The position of the kth individual at time t is given by
We define the empirical measure of the system by:
Step 2. Now we consider R different classes of particles and we denote by the number of particles in class i, . The equations of movement for each particle are as above. In addition, we assume that each particle of the class i can
- • die, at a rate given by , where P denotes the position of the particle;
- • give birth to a new particle of the class q, at a rate given by ;
- • pass to a different class particle, at a rate given by , for , with .
We define an empirical measure
4.2 SDEs of the IBM
We start with a few words about what we mean by a Poisson process with variable intensity [30]. Let be an integer-valued stochastic process defined for every time . By saying is distributed as a Poisson process with variable intensity we mean its probability density is given by
Combining the motion and the demographic processes:Remark
We assume, for sake of simplicity, that each integer number k is used only once throughout the simulation; this means any newborn particle will be assigned an unused index k, which will not be used again after its death. As such, each particle k will have a unique lifetime interval. The convenience of this assumption is notation-related and will be apparent in subsequent paragraphs. In the next section we will explain the SDEs which describe the evolution of the four types of particles considered in the IBM, in terms of the corresponding Poisson processes.
4.2.1 Cells
- •
Motion. The position and age of the kth cell at time t are represented by where is the horizontal position, is the vertical position, and is the age. According to the explanation given before in Section 3.1.1, we have:
where are independent Brownian motions for each cell k. The components represent the haptotaxis, i.e., a displacement proportional to the gradient of MM concentration, while the third component is the aging speed, equal to one per time unit. is a term storing the intensity of the Brownian motion, which increases with the mutation type. - • Demography. The model includes the following Poisson processes:
4.2.2 MM
- • Motion. MM particles experience no motion, so their position remains fixed throughout the simulation and neither transport nor Brownian effects appear.
- • Demography. Only one death Poisson process appears: degradation at a rate given by ηm (see Section 3.1.2).
4.2.3 MDE
- •
Motion. MDE particles experience Brownian motion, so the motion of the kth particle is given by
where stands for the diffusion coefficient and are independent Brownian motions for each MDE particle k. - •
Demography. We consider the following Poisson processes:
- – Production: MDE particles are produced by cells at a rate given by (4);
- – Decay: MDE particles disappear at a constant rate .
4.2.4 Oxygen
- •
Motion. Oxygen particles experience Brownian motion given by
- •
Demography. We consider the following Poisson processes (see Section 3.1.4):
- – Production: oxygen particles are produced by MM particles at constant rate .
- – Uptake: oxygen particles are uptaken by cells of type i at a rate .
- – Decay: oxygen particles disappear naturally at a constant rate .
4.3 Obtaining the PDE system for the IBM
As explained in Section 4.1, we develop the following limiting PDE system for the IBM:
- • Cells.
- – Proliferating state:
- – Quiescent state:
- – Boundary conditions:
. The boundary conditions appear naturally from the fact that a proliferating cell gives birth to two daughters, whose mutation type can be either the same (in case no recent mutation has occurred) or the next sequential type (in case the last division led to a new mutation). In our simulation we have taken as in [16].
- – Proliferating state:
- • MM.
- • MDE.
- • Oxygen.
5 Numerical analysis of the PDE
To analyze this model computationally we used a finite difference discretization in space that for the range of parameters of this work, allows large time steps. For the 2D version, we use and , in the domain .
The results of the simulations can be seen in the figures. The interested reader can also check some videos (.avi format) of the numerical resolution on http://dmaii.etsii.upm.es/~cells.
A fully implicit scheme is far too expensive for practical calculations. We succeeded in using an improved predictor-corrector scheme with a double purpose: to test the convergence of our straightforward explicit discretization in time and space and to develop a robust general method for accurately solving the system given in Section 4.3. Discretization of the age variable does not suffer the difficulties mentioned in [17] because our methods are not semi-discrete.
In order to obtain a more complete picture of the process, we have started extending the two-dimensional work to three space dimensions. The experiments reveal that the general morphology is similar to that of the 2D version. As a significative (though preliminary) example, Fig. 8 shows a 3D shell of type-4 cells, with an inner necrotic core represented as an area void of cells. In the 3D case, we have been able to efficiently compute different tumors with and . Moreover, the finite-difference procedures are well-suited for parallel implementation.

A picture of a 3D simulation of the IBM. The inner core is quiescent and necrotic.
We are presently developing a splitting procedure and different approximations of the div-grad operator to allow the accurate modeling of morphologic instabilities, and in particular, more intricate and irregular tumor boundary evolution and satellite tumors around the main tumor mass.
6 Conclusion
In this work we have studied the invasion of tumor cells into surrounding tissue by using two approaches: one, a computational individual-based perspective; the other, a partial differential equations perspective reproducing the same phenomena. Two computer experiments were carried out for both approaches: (a) one starting with an initially homogeneous MM density (homogeneous case); and the other (b) starting with an initially heterogeneous MM density (heterogeneous case). We considered four types of cell phenotypes evolved by sequential mutation from the wild-type. The higher the mutation type in the sequence, the higher the cell's aggressiveness in the invasion.
Simulations show the expansion of increasingly aggressive cell phenotypes, as can be observed in Figs. 2 and 3. In the homogeneous MM case, cells evolve in a ring-shaped fashion (Fig. 2). We note that quiescent and necrotic cells are not drawn in the figures and the inner zone of the ring contains only quiescent and necrotic cells. Proliferating cells advance outwards, attracted by MM. In the heterogeneous case, cells do not form a ring, but advance toward higher concentration of MM (Fig. 3).

Snapshots of the evolution of the IBM (homogeneous MM initial distribution). For clarity of representation, quiescent cells are not represented. The color indicates the mutation (blue = wild-type, cyan = type 1, green = type 2, orange = type 3, red = type 4).
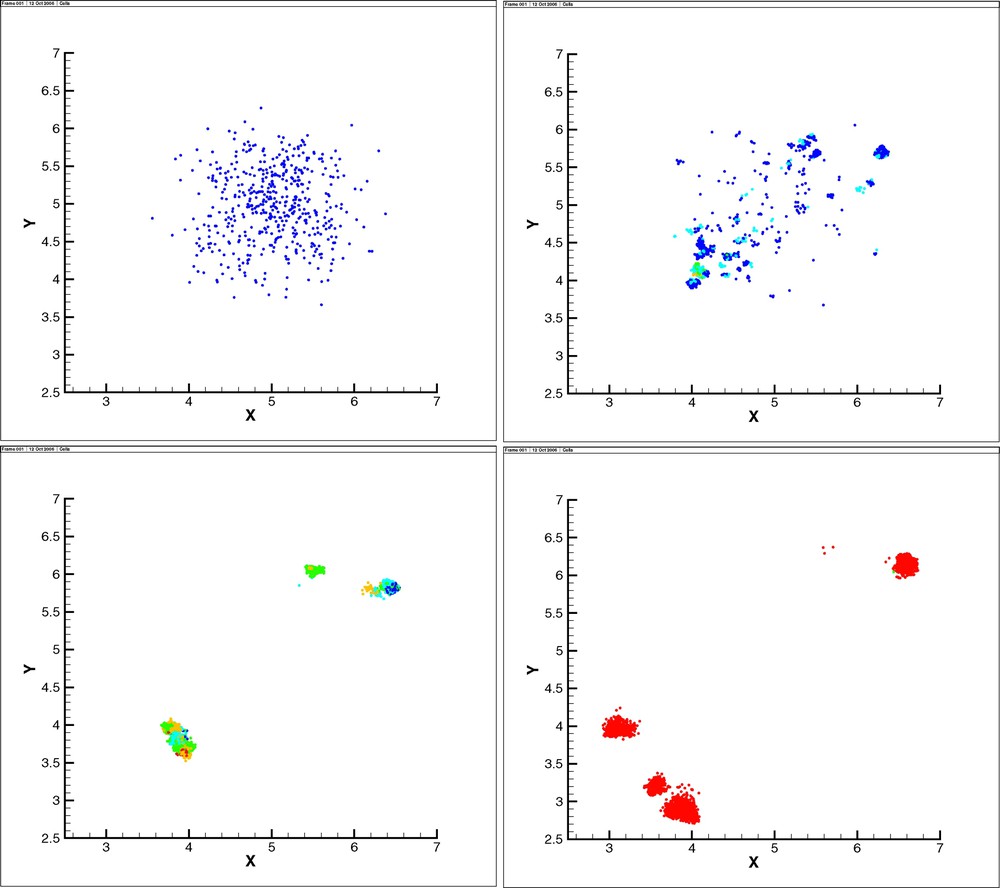
Snapshots of the evolution of the IBM (heterogeneous MM initial distribution). As in Fig. 2, for the sake of clarity, only proliferating cells are represented. The color indicates the mutation with the same code (blue = wild-type, cyan = type-1, green = type-2, orange = type-3, red = type-4).
An important biological feature of tumor invasion is fingering patterns (Fig. 7). Our simulations systematically reproduce such patterns, both in the homogeneous case (Fig. 2) and the heterogeneous case (Fig. 3). Fingering was less evident in the homogeneous case, nevertheless it appears even under fairly uniform densities of MM. We conclude that very slight oscillations of MM density may be enough to induce finger patterns. We hypothesize that in vivo MM concentrations may present slight density heterogeneities, which are eventually responsible for fingering patterns.
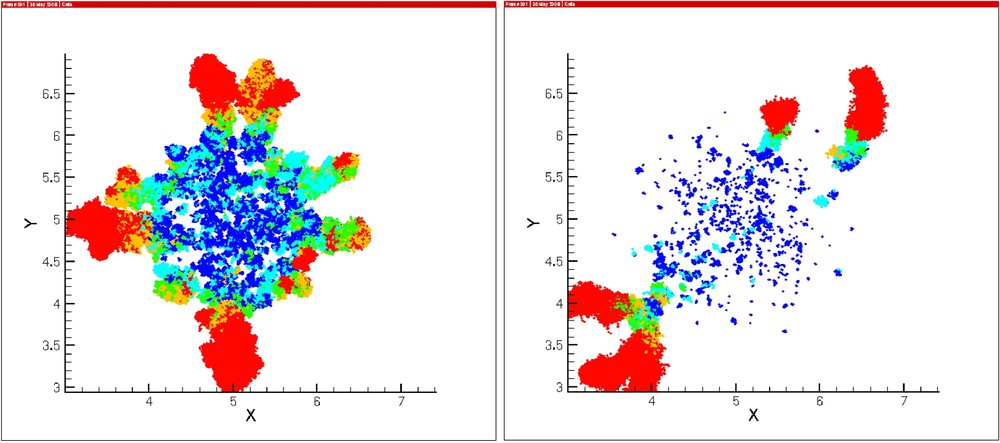
Finger patterns appear due to MM density fluctuations, not only in the heterogeneous case (right), but in the homogeneous case (left) as well.
Our simulations are the results of spatially explicit, fully continuous, individual based computation, whose building blocks are tokens for cells, MM, MDE elements and oxygen particles (Fig. 4). We derived an equivalent (in a mathematical sense) PDE model to describe the same behavior. The PDE numerical simulations match remarkably well those of the IBM. The homogeneous case develops a crown-shaped cell density (Fig. 5), with inner zone density close to zero, as corresponding to the quiescent and necrotic zone. In the heterogeneous case (Fig. 6), two main directions of invasion appear, as a consequence of the initial spatial configuration of the MM density. The concordance of the IBM and PDE approaches is evident in Figs. 2 and 5 and Figs. 3 and 6.
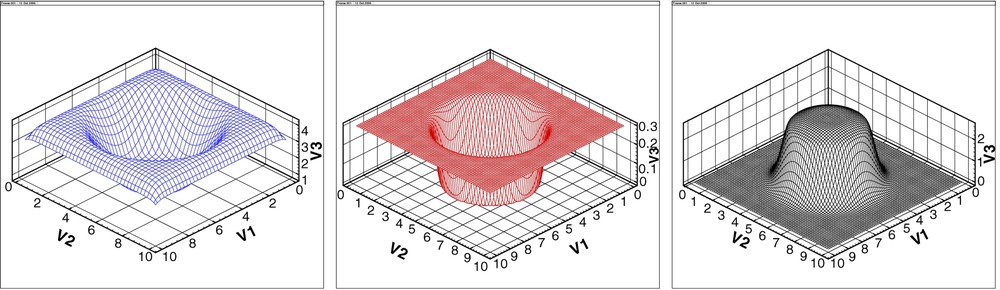
The three boxes represent the oxygen, MM, and MDE density profile, respectively, at a representative stage of the simulation.

Snapshots of the type-4 cells density evolution, as provided by the numerical solution of the PDE (case of homogeneous MM initial distribution). Only type-4 cells are taken into account in order to make the figure interpretation easier.

Snapshots of the type-4 cells density evolution, as provided by the numerical solution of the PDE (case of heterogeneous MM initial distribution). As in Fig. 5, only the most aggressive (type-4) cell density is represented.
IBM computational simulations are accessible and intuitive. They do not require deep mathematical knowledge and can be very helpful for non-experts. However, high computation costs appear when dealing with many particles; in particular, the computation of interactions between N particles increases (roughly) with , with making such simulations hardly feasible for very large values of N. As a consequence, it is important to acquire a correspondent PDE approach that reliably reproduces equivalent – at least in the mathematical sense of Section 4 – results. We believe IBM modeling to be valuable, but its practical limitations must be known and appreciated.
From a paradigmatic point of view, it seems to us more natural to model large tumor cell populations as continuous mathematical densities rather than as frameworks of discrete units, even taking into account the variable nature of individual cells. Furthermore, we believe the continuous approach in an IBM overcomes some limitations of the discrete one, bridging the gap between many-particle systems and differential equations-based models. We find it particularly relevant and fruitful to establish comparisons between computational and PDE approaches. Not only can such comparisons give insight into complex population behavior in general (not only tumors), but can as well be tools to test models. The scheme in Section 4 to establish the comparison between IBMs and PDEs remains valid in general cases, and can be used by computational modelers seeking mathematical validation.
As for future work, we plan to extend our work to three dimensions. In fact, we have already run tentative 3D simulations, which showed similar results to the 2D case. Fig. 8 is a snapshot of a 3D simulation displaying a quasi-spherical tumor with a necrotic inner core. Fingering patterns were also observed in our 3D simulations, which are still under development.


