1 Introduction: The biology of Buchnera aphidicola
1.1 From Paul Buchner to Hajime Ishikawa
Symbioses between insects and bacteria are widespread in the biodiversity. Aphid symbiosis with specific intracellular bacteria was first described in detail by Paul Buchner in his famous book “Endosymbiosis of animals with plant microorganisms” [1]. At the same time, P. Buchner also contributed to developing the concept of symbiosis within the scientific community, and he formulated the nutritional hypothesis for obligate intracellular endosymbionts in insects “endosymbiosis must in some way be caused ecologically, their occurrence is usually linked with unbalanced feeding”. In fact, Buchnera aphidicola (the name of the bacterium was proposed by Paul Baumann's team [2,3]), is associated with most phloem sap feeding aphids, i.e. it is not present in Phylloxeroideae, but is present in all Aphidoideae, except the Cerataphidini tribe in which Buchnera1 has been replaced by specific symbiotic yeasts [4]. Buchnera is closely associated with the life style, and as a consequence with the physiology of aphids. Buchnera provides the essential amino acids that the aphid cannot find in its peculiar diet consisting solely of phloem sap. Symbiosis with Buchnera is an important factor in making the aphids major pests. Nowadays, Buchnera is one of the most studied obligate intracellular bacteria of insects [5]. Thirty-five years after Paul Buchner's book, the first genome sequence of the Buchnera from the pea aphid Acyrthosiphon pisum (BAp) was completed by the team of Hajime Ishikawa at the Riken Institute in Japan [6]. Three other complete sequences of Buchnera from divergent aphid species are now also available: BSg from the cereal aphid Schizaphis graminum [7], BBp from the pistachio-tree aphid Baizongia pistaciae [8], and BCc from the cedar aphid Cinara cedri [9]. Recently, seven more BAp genomes have been sequenced using the Solexa sequencing technique [10], making Buchnera one of the most popular models in symbiotic bacterial evolutionary research.
In this review, after briefly describing what is currently known about the biology of Buchnera (and particularly that of BAp), we focus on the structural aspects of its genome, and on its genetic and metabolic networks with the aim of providing a systemic description of the symbiotic function of the aphid endosymbionts.
1.2 Phylogenetics, cospeciation and origin of symbiosis
The phylogenetic positioning of Buchnera is still ambiguous because of the high evolutionary rate and strong AT bias of its genome (that affects most of its genes). Nevertheless, several evolutionary studies have been performed, and they have tended to converge toward positioning Buchnera within the group of the γ3-Proteobacteria [2,3,11,12]. Hence, Buchnera is closely related to the Enterobacteriaceae including Escherichia coli. The name Buchnera aphidicola has been attributed to all the lineages of aphid endosymbionts using BSg, the endosymbionts of the wheat aphid Schizaphis graminum, as the type species [3]. Since Buchnera is strictly vertically transmitted through aphid generations, speciation of host lineages has been accompanied by the divergence of the associated endosymbiont lineages, resulting in perfect congruence between the phylogenetic trees of the two groups of species. Divergence dates have been estimated on the basis of aphid fossil records and gene evolution rates [13] in order to trace speciation events within the Buchnera clade, and compare the evolutionary rates of different lineages [10,14]. The association has been estimated to have begun around 150 Myr ago [13,15]. During this co-evolution process both partners have become completely interdependent: when aphids are deprived of their primary symbiont, they are no longer able to reproduce, and so far it has been impossible to culture Buchnera without its host in vitro.
1.3 Localization of Buchnera within the aphids
Buchnera is located within specialized eukaryotic host cells, known as bacteriocytes, which are located inside the aphid body cavity, adjacent to the ovariole cluster. The bacteriocytes (about 60 to 80 in each aphid) are giant polyploid cells within which Buchnera resides in the cytoplasm surrounded by a host-derived membrane, the symbiosome (Fig. 1). The bacteriocytes are assembled in a bilobed organ-like structure known as the bacteriome. In embryos, a thin layer of syncytial cells surrounds the bacteriome, whereas in old aphids the bacteriome structure degenerates in parallel with the nutritional and reproductive decay of the insect [5].
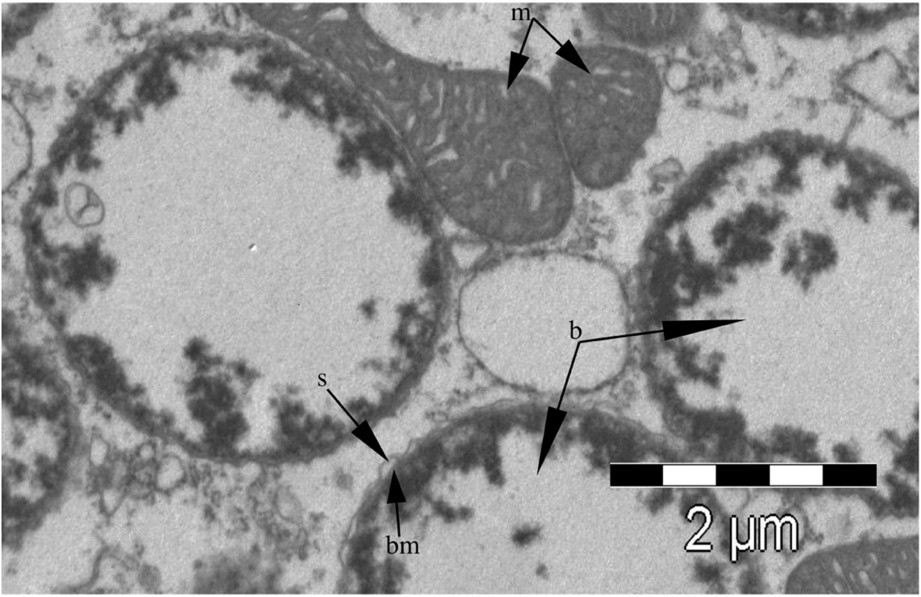
Scanning electron microscopy of Buchnera aphidicola within a bacteriocyte from the parthenogenetic female of Acyrthosiphon pisum. Several features are visible: s – symbiosome membrane, bm – cell wall of Buchnera, b – Buchnera within the symbiosome, m – mitochondrion.
The most active state of development and reproduction of the aphids is during the cycles of parthenogenesis when the wingless aphid mothers produce living larvae. During this period of telescoping generations (i.e. the larvae include embryos in their own body), two distinct populations of Buchnera coexist inside the aphids: the maternal and embryonic populations. The embryonic Buchnera population corresponds to about 75% of the total Buchnera population of the mother [16]. The growth rate of Buchnera seems to reach its peak during embryo development, just after the young embryos have been colonized [17], whereas the number of Buchnera remains stable in adult aphids, and declines as the aphids get older [5], suggesting a slow growth rate of the bacterium within maternal bacteriocytes.
1.4 Secondary endosymbionts in aphids
Like most eukaryotes, aphids can be considered to be complex micro-ecosystems involving consortia of viruses and bacteria, interacting in communities. Part of this diversity occurs in the flora of the digestive tract [18], but also inherited intracellular symbionts are also highly diversified in aphids [19]. In addition to Buchnera, most natural aphid populations also harbor other intracellular symbiotic bacteria, known as secondary endosymbionts. In general, these non-obligatory, secondary endosymbionts are not present in all aphid species nor in all the individuals of the same population. They are transmitted vertically, although horizontal transfers may occur between different aphid populations and species. These bacteria are fairly diverse, and include species of the genera Spiroplasma, Wolbachia, and Rickettsia, as well as several Enterobacteria (Regiella insecticola, Hamiltonella defensa, Serratia symbiotica). Secondary endosymbionts have been shown to have the following functional roles: thermal resistance [20], adaptation to the host plant [21], resistance to parasitism [22] and complementation of the nutritional role of Buchnera [9]. Hamiltonella defensa can invade some bacteriocytes usually reserved to Buchnera [23], and has been shown to harbor active bacteriophages, known as APSE-1 and 2, which encode toxins that may be involved in invading host cells or, on the contrary; protecting them against parasites [24]. To reach a better understanding of symbiosis and of the evolution of Buchnera, these secondary endosymbionts have to be taken into account; some recent experimental results suggest that in some laboratory aphid strains other bacteria may replace Buchnera [25].
1.5 Transmission of Buchnera and population dynamics
Buchnera is transmitted vertically through the host generations, and each host generation has to undergo an infection phase. The infection of embryos with bacteria from the mother occurs during the blastoderm-stage via an opening in the posterior pole of the embryo in the viviparous morphs, whereas in the oviparous morphs, it is the eggs that are contaminated [1,17,26]. One direct consequence of this infection phase is that the symbiont population passes through successive ‘bottlenecks’, i.e. only a small proportion of the maternal symbiont population is transmitted to the offspring. Wilkinson et al. have shown that a single maternal bacteriocyte is the source of all the Buchnera cells in a given embryo, and that Buchnera enters the embryo via a specific membranous duct [17].
Hence, Buchnera populations are of small size, suffer drastic bottlenecks and are isolated within the bacteriocytes, thus preventing any possibility of any exchange of genetic material occurring between bacterial lineages. This peculiar pattern of population dynamics increases the probability that mildly deleterious mutations will become fixed, and also makes it impossible for them to be corrected (this is known as Muller's ratchet effect). Indeed, Buchnera is subjected to increased genetic drift that imposes a bias in genome composition and accelerates evolutionary rates even at non-synonymous sites [14]. This acceleration of the evolutionary rate also reflects the effect of higher mutation rates (due to mutator genes and/or the absence of a repair system), as well as changes in selection pressure due to the intracellular environment [10,14,27–30].
1.6 Interaction with host physiology
Aphids have developed specialized mouthparts featuring long flexible stylets that they insert into the plant tissues until they reach the phloem cells, from which they exude the sap. This phloem sap constitutes the sole food source in the aphid diet. It is rich in carbohydrates, but deficient in nitrogenous compounds, and so constitutes a very unbalanced diet, and one with a composition that varies depending on the plant, and also on the biotic and abiotic environmental conditions for the same plant. Like those of other insects, the nutritional needs of aphids also vary with their stage of development.
The provision of aphids with nutrients by Buchnera has been extensively discussed, and is the subject of considerable speculation [5]. Experimental studies using a combination of controlled artificial diets and antibiotic-treated aphids (aposymbiotic aphids), as well as genomic information, have focused on nitrogenous compounds that are not present in the phloem sap, and especially on essential amino acids. Douglas et al. have provided evidence for the synthesis of methionine by Buchnera in the aphid Myzus persicae [31], and that of tryptophan in BAp [32]. The involvement of Buchnera in the transfer of nitrogen from glutamine or glutamate has been demonstrated by Sasaki and Ishikawa [33]. Febvay et al. have provided direct evidence for the biosynthesis of threonine, isoleucine, leucine, valine and phenylalanine by BAp [34,35], highlighting the ability of Buchnera to adapt its production of amino acids to the aphid's nutritional needs [36].
If at present we know the qualitative metabolic contribution of Buchnera to the symbiotic interaction [37], the quantitative contribution of Buchnera to the adaptability of the variation of the aphid nutrition remains to be deciphered. Buchnera is often described in the literature as a degenerating bacterium, with low sensitivity to external conditions, and an inability to regulate its transcriptional and post-transcriptional responses to the variable demands of its host [38]. New insights into the symbiotic function of Buchnera provided by systemic analyses have been made possible by the fact that several complete genomes of Buchnera and other endosymbionts are now available, as well as specific tools developed for network analyses.
2 Genomic analysis of Buchnera
2.1 Structural properties of the Buchnera genome
The genomes of the different strains of Buchnera, ranging in size from 420 to 650 kb, are among the smallest of the prokaryotic genomes to be sequenced so far [39]. The process of genome shrinkage in Buchnera has been extensively studied. Initial studies determined the gene content of the last common ancestor (LCA) Buchnera with its close relative E. coli [6,40,41]. Van Ham et al. [8] and Silva et al. [42] subsequently estimated the genome content of the last common symbiotic ancestor (LCSA) of the four sequenced strains in order to reconstruct the process of gene loss in each lineage (reviewed in [43]). Two models of gene loss have been proposed in order to explain the extreme genome shrinkage of Buchnera. The first model, proposed by Moran and Mira [41], distinguishes between the early massive gene losses caused by the fixation of large deletions and the relatively minor gene losses that have followed as a result of the pseudogenization process. According to this model, the process of genome shrinkage was highly dynamic during the early stages of symbiosis (during the transition from the LCA to the LCSA), resulting in the removal of large genomic areas containing Buchnera genes that had become redundant or unused in the context of the stable intracellular environment [40,41]. The second model, proposed by Silva et al. [40], attributes the genome shrinkage to multiple gene disintegration events dispersed over the whole genome. Recent comparative analyses of the sequenced endosymbiont genomes support this second model [44–46].
It has been proposed that in some Buchnera lineages this dynamic step was followed by a long period of stasis, lasting for more than 50 Myr, due to the loss of insertion sequences and of the genetic elements that mediate an efficient homologous recombination, as well as the reaching of an equilibrium between the number of genes and the symbiosis function [7]. However, the extreme reduction found in other lineages, such as in BCc [9], as well as those revealed by the recent work of Moran et al. [10] on BAp, have revealed that deletions and inactivations eroding the genome of Buchnera are ongoing and do not appear to have reached a limit.
The genome of BAp is composed of a 640 kb circular chromosome plus two plasmids, pLeu and pTrp, carrying all the genes specifically required for the leucine biosynthesis, and two genes encoding the enzymes for the tryptophan biosynthesis pathway [47,48]. The sequence analysis of the two plasmids suggests that they have different replication mechanisms and independent origins [43,49]. However, more accurate analyses are needed to confirm this later hypothesis. Plasmid-borne copies were acquired from an ancestral Buchnera genome, and not by horizontal transfer from an external source [49]. The evolutionary history of the two plasmids reveals highly dynamic recombination process, including an original transfer from the chromosome to the plasmids, and four independent back-transfers and gene-order reorganizations [43].
It is important to note that the genome of Buchnera is highly polyploid [50]. Buchnera has large cells about 3 to 5 μm in diameter (about 15× the volume of one E. coli cell), and contains 10× more DNA than E. coli [50]. Indeed, incomplete division due to an ineffective replication and segregation system may be the cause of the polyploidy in Buchnera [7]. The chromosome copy number per Buchnera cell varies from 50 to 200, depending on the age of the aphids [50,51]. Variable relative numbers of plasmid and chromosome copies within a Buchnera cell were probably responsible for the leucine and tryptophan gene transfers and back-transfers that have occurred between plasmids and chromosome during the evolution of Buchnera [43]. This dynamics of copy number could also be a mechanism of regulating gene expression; however, to date no experimental evidence has been found to support this hypothesis.
The genome of Buchnera consists of 70% of AT bases (90% in the intergenic regions). Important composition biases between the two DNA-strands are observed, and they make it possible to localize both the origin and the terminus of replication of the bacterial chromosome (Fig. 2). The genome of Buchnera is tightly packed with genes (some with overlapping coding sequences), indicating that superfluous DNA was rapidly eliminated during evolution [52,53].
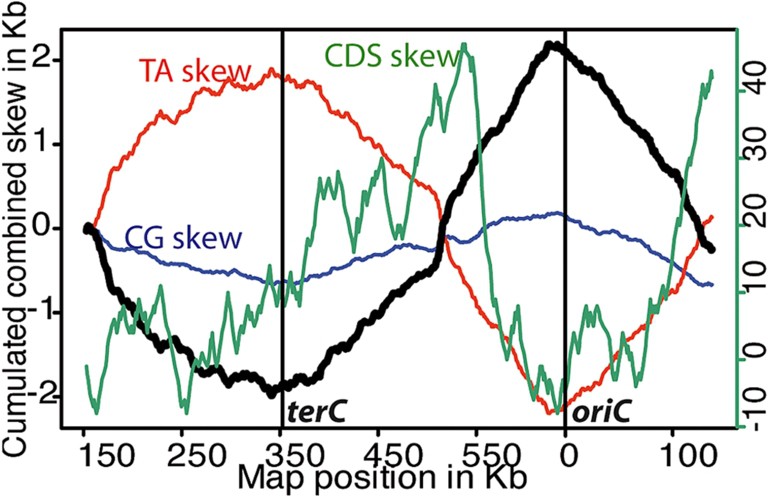
Scan of the gene and base composition of the Buchnera aphidicola Ap genome obtained with Oriloc [85]. Green CDS skew (right scale): coding sequence orientation bias, when the coding sequence is on the forward strand, the cumulative CDS skew is incremented by one (and conversely when it is on the reverse strand, the cumulative CDS skew is decreased by one); red TA skew (right scale): absolute T–A skew in the 3rd codon position of the cds; blue CG skew (right scale): absolute C–G skew in the 3rd codon position of the cds; black cumulated skew (left scale): points the replication origin (the positive peak), and the replication termination (negative peak).
Gene organization along the Buchnera chromosome is shown in Fig. 3. A gene distribution bias between the leading and lagging DNA strands can be seen: 56% of genes are encoded by the leading strand. The bias becomes even more marked if we consider only the essential genes: 60% of the essential genes are located on the leading strand. We investigated the effect of gene DNA strand position, operon organization and gene essentiality on the variability of the basal mRNA abundances in BAp: putative transcription units (ANOVA, , ) and gene essentiality (ANOVA, , ) are both significant factors in this model. No significant effect was observed between strand position and gene expressiveness, indicating the absence (or the low) selection for gene expression in Buchnera when the genome is replicating [54]. This finding is consistent with the slow growth rate of Buchnera.
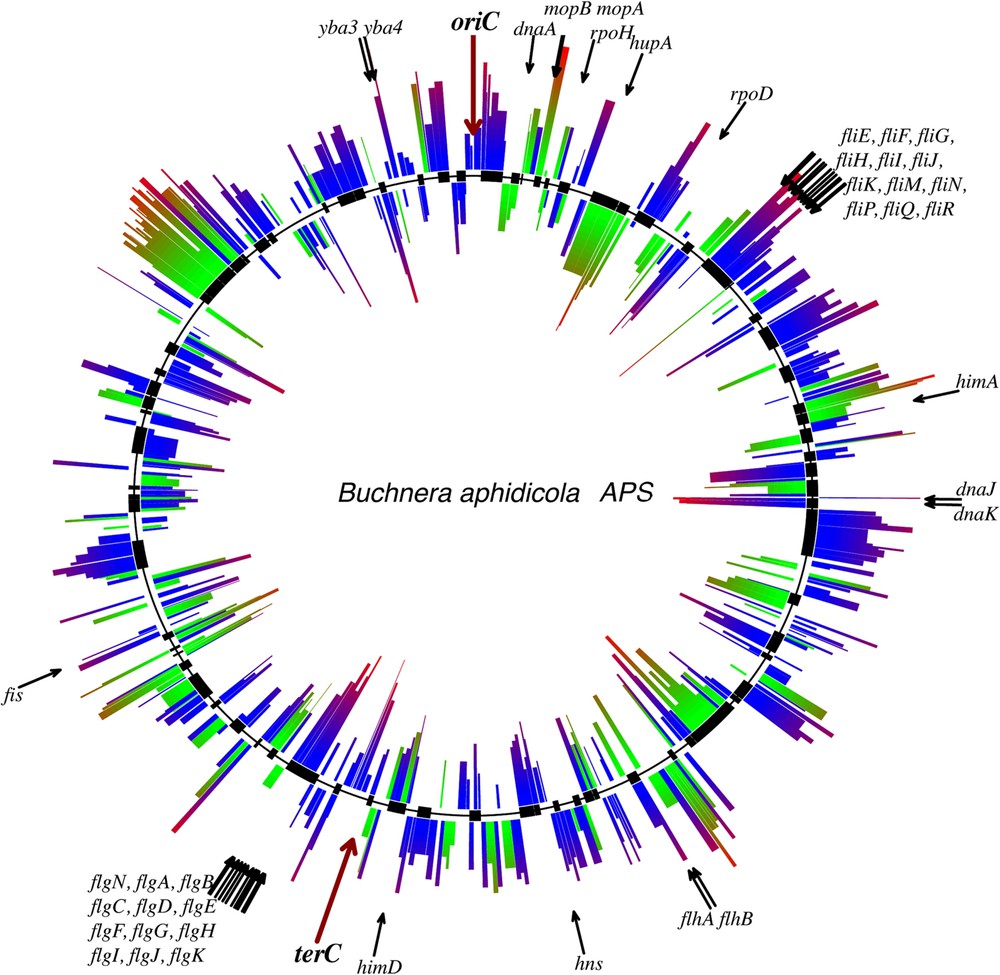
The chromosome architecture of Buchnera aphidicola Ap. Graphical elements outside (resp. inside) the middle circle characterize the leading (resp. lagging) strand genes. The height of the segment associated with each gene indicates the gDNA-normalized expression level [54]. Gene expression is also indicated by the color gradient: from green (blue) for a low level expression, to red for a high level expression of essential (non-essential) genes. The essential genes in Buchnera aphidicola Ap were those proposed by Gil et al. [86]. Black rectangles on the middle circle identify putative operons annotated using synteny analysis of the BAp chromosome and of E. coli.
The genes in Buchnera are organized into putative transcription units (PTUs) that have been inferred from their orthology and synteny with E. coli (Fig. 3). No specific regulators corresponding to these operon structures have been preserved in Buchnera (see below). However, experimental data using dedicated microarrays reveal a significant correlation between basal and differential gene expression (following nutritional stress applied to the host aphids) and PTU organization, suggesting that these operon structures play a functional role in regulating gene expression in BAp [54,55].
2.2 Gene expression, transcription regulation and the genetic network of Buchnera
Genomic analysis has revealed that there are virtually no genes for regulatory systems in the BAp genome [6]. Indeed, two-component regulatory systems are also absent. None of the E. coli orthologous regulators of the operons encoding the enzymes of the essential amino acids pathways are present in Buchnera. The genes of Buchnera have no leader sequences, and the bacterium does not use attenuation systems [6]. The genes encoding adenylate cyclase (cyaA) and the AMPc receptor (crp) are absent in BAp, indicating the absence of an ability to respond to a change in carbon source, a system that is usually highly conserved in prokaryotes. Only two sigma factors are present in BAp: rpoD and rpoH. Buchnera does not display any clearly defined transcriptional terminators, and the binding sites for the generalized sigma-70 promoter, required to initiate the transcription of most genes, are not readily distinguishable. However, BAp has retained genome ancestral regulators that may control global shifts in gene expression, corresponding to different stages of the host's life cycle. This global change may be sensed by the accessibility to certain precursors (i.e. ATP), and the effectors may be Nucleoid Associated Proteins (NAPs), such as IHF and FIS. Regulation via cell division and/or genome ploidy (which alters the likelihood of interactions between transcriptional regulators and gene promoters) may also be involved in these gene expression shifts.
Several experiments have been performed to analyze gene expression in Buchnera. PCR fragment-based microarrays [38,56,57] and oligonucleotide microarrays have been produced [54,55,58,59], physiological experiments were conducted, and evidence has accumulated revealing weak gene transcriptional regulation. Gene expression normalization using genomic DNA (gDNA) has been used to compare the gene expression level between genes under the same experimental conditions [54]. Using this approach, Viñuelas et al. [54] experimentally defined a set of highly expressed genes for BAp on the basis of the gene GC rate, which were divided into two groups of slowly and quickly evolving genes respectively (some of them are shown in Fig. 3).
The group of conserved and highly expressed genes contains most of the ribosomal (not shown in Fig. 3) and chaperone (dnaK, dnaJ and mopA, ipbA) protein genes. Moran et al. [38,60,61] have provided evidence that most of the 20 heat shock genes retained in the Buchnera genome are constitutively over-expressed compared to other bacteria. Several studies have been performed on the GroEL protein, originally called symbionin, as it is the most abundant protein of Buchnera [62] representing almost 10% of the total amount of aphid proteins found in BSg [63]. It is supposed that GroEL and the other chaperonins buffer the effects of translation errors so that the proteins that have accumulated mildly deleterious mutations in Buchnera can still fold correctly [14,64].
Most of the flagellar genes (11/16), and the two orphan genes of Buchnera (yba3, yba4) have been identified among the fast-evolving highly expressed genes [54]. This observation is particularly intriguing, since Buchnera is a non-motile bacterium which has reduced its genome by eliminating energetically expensive, unnecessary or redundant genes [27]. A consensus hypothesis is that flagellar genes are in fact involved in an essential symbiotic function, the export of protein to the host [6,54,65], as is known to be the case in Salmonella typhimurium and Yersinia enterocolitica [66,67]. The high level of expression of most of these genes, their amino acid sequence divergence and the particular subset of flagellar genes conserved among the 4 sequenced Buchnera strains preserving only a minimal type-III virulence secretion system, support this hypothesis [54].
In most bacteria, selection for gene expressiveness is reflected in codon usage. Codon usage in Buchnera is dominated by an AT bias. Indeed most preferred codons end with A and T in Buchnera [68]. However, a significant correlation between tRNA-isoacceptor abundances and codon usage has been shown in Buchnera, highlighting selection for gene expressiveness [59]. Codon usage was observed thanks to the true set (measured experimentally) of highly expressed genes in Buchnera. Genes that are highly expressed in Buchnera use more C-ending rare codons, and fewer G-ending codons than more weakly expressed genes (Fig. 4). This tends to favor optimal codons for some specific amino acids in the highly expressed genes of Buchnera, suggesting the existence of a qualitative selection of the translation of certain sites (amino acids) that are functionally important in some of the essential proteins of Buchnera [59]. Recent work by Toft and Fares (2009) [69] has confirmed this hypothesis.

Codon usage in Buchnera aphidicola Ap. The height of the bars corresponds to the differential ratio of RSCU (relative synonymous codon usage) between the most and least expressed genes in Buchnera (defined in Viñuelas et al. [54]). Codons are grouped by amino acid. Third-codon bases are indicated at the top of each bar. Asterisks are used to indicate amino acids with significant differences in codon effectives (Chi-squared test, α = 0.05).
A global shift of gene expression in bacteria is usually governed by a change in DNA topology [70]. In Buchnera, the rates of gene expression along the chromosome were analyzed using autocorrelation models and spectral analysis [54]. Significant periodic components in the gene transcription levels along the chromosome of the BAp genome have been identified: small- (2 to 20 genes), medium- (20 to 70 genes) and long-range (80 to 152 genes), and such periodic signals have been observed in E. coli and Bacillus subtilis [71,72]. In order to determine the role of operons in such periodic structures, gene permutations were introduced in silico into the Buchnera genome. Four simulated permutations were defined as follows: (1) disrupting the gene order between the operons; (2) disrupting the gene order between the operons as well as the number of singletons between them; (3) disrupting the gene order between the operons as well as the operon ranking; and (4) disrupting the order of all the genes on the chromosome. The results obtained after these permutations suggest that operon organization in BAp is partly responsible for small periodic signals and, in addition, operon spacing and ranking contribute to the medium and long-range periods, suggesting that the BAp chromosome has a periodic topological structure [54].
Finally, a global analysis of the putative transcriptional regulators conserved in Buchnera was carried out in order to reconstruct the genetic network of the bacterium. Eight proteins, orthologous with the transcription factors of E. coli, were identified in Buchnera. This demonstrates that the genetic network of Buchnera has undergone extreme shrinkage (Fig. 5) conserving generic regulators that are probably only sensitive to major stress within the cell, where specific tuning is no longer necessary. In fact, five of the conserved transcription regulators belong to the NAP family (Nucleoid Associated Protein): (1) H-NS, histone-like nucleoid structuring protein; (2) HU, heat-unstable nucleoid protein; (3) IHF, integration host factor; (4) DnaA, DNA-binding protein A; and (5) FIS, factor for inversion stimulation. Bioinformatic structural analysis (unpublished data) reveals that these five NAPs display high levels of conservation of important amino acids and functional domains, indicating that this function has undergone severe purifying selection.

The genetic network of Buchnera aphidicola Ap. This network was obtained from the RegulonDB genetic network of E. coli (http://regulondb.ccg.unam.mx/), by conserving only the regulator genes of Escherichia coli (and their corresponding targets) which have an orthologous gene in Buchnera. This first rough approach highlights the centrality of the two main players, FIS and IHF.
The conservation of a significant proportion of the NAPs set commonly found in bacteria (including both specific and non-specific-DNA binding NAPs) offers an especially intriguing potential explanation for gene expression shifts in BAp via DNA topology variations. This is that the combined activities of NAPs and topoisomerases shape DNA topology, which is known to act like a transcription factor in regulating gene expression [70]. NAPs are also involved in the organization of chromosome macro-domains via their involvement in the formation of a dynamic topological barrier [73], and so may contribute to the medium- and long-range periodic gene expression profiles observed in BAp.
3 The gene repertoire and the metabolic network of Buchnera
The initial genetic research on Buchnera was performed by the team of Paul Baumann on BSg (reviewed in [5]). They studied the genes in the synthesis pathways of several essential amino acids, highlighting gene conservation and synteny with the genome of E. coli, and hypothesized that genome reduction had occurred. Analysis of the complete sequence of BAp by Shigenobu et al. [6] revealed a limited gene repertoire, and offered a first global overview of the metabolic network of the bacterium.
The housekeeping system is minimal in Buchnera. It has a limited capacity for DNA repair and recombination, and lacks the genes essential for recombination in bacteria, the recA and uvrABC genes required for the excision repair system [41], and the SOS system genes. Genes coding for the methylation proteins and for the restriction enzymes are also missing. The translation machinery is complete (although minimal) with 32 tRNA, 55 ribosomal proteins, 12 translation factors and 21 tRNA amino-acyl transferases [59]. It is noticeable that rRNA genes are present as single copies in Buchnera, whereas 7 copies are present in E. coli. This is generally considered to be a characteristic of slow-growing organisms.
The analysis of metabolic genes reveals that Buchnera has preferentially retained genes coding for multipotent proteins (unpublished data). The central metabolism in Buchnera is based on the use of sugar (glucose or mannitol) via the glycolysis and pentose phosphate pathways. However, Buchnera lacks a complete TCA cycle. Carbon and nitrogen fluxes are mainly oriented towards the biosynthesis of essential amino acids. Buchnera has retained the enzyme repertoire necessary for the carbon backbone biosynthesis of arginine, threonine, lysine, valine, isoleucine, leucine, tryptophan, phenylalanine and histidine. These 9 essential amino acids are synthesized by Buchnera via standard bacterial pathways, from precursors including some non-essential amino acids, plus α-ketoglutarate, 3-phosphoglycerate, oxaloacetate, pyruvate, phosphoenol pyruvate, 4-erythrose-phosphate and/or ribose-5-phosphate imported from the host cell or synthesized starting from the catabolism of sugars. Buchnera has also conserved a complete gene set for the sulfur reduction pathway, and has retained the ability to biosynthesize cysteine, presumably using serine and adenosine-5-phosphosulfate (APS) as precursors. The biosynthesis of methionine by Buchnera is, however, carried out only using homocysteine supplied by the host as precursor.
Buchnera breathes aerobically, and lacks the genes for fermentation and anaerobic respiration. Buchnera is able to regenerate ATP from ADP via a functional ATP synthase F0F1 system. It is however not clear whether Buchnera has conserved the capacity to synthesize all its nucleotides. Indeed, the bacterium does not posses all the enzymes encoded by E. coli that are required for the interconversion of nucleotides between their mono-, di-, and tri-phosphorylated and deoxy forms. The Buchnera enzymes may display broader specificity or, alternatively, essential precursors may be imported directly from the host cell [74].
Buchnera has retained the complete gene set required for the biosynthesis of lipoic acid, FAD (from neosynthesized riboflavin) and NAD (from bacteriocyte-imported nicotinate), as well as the terminal enzyme gene set for biotin biosynthesis. Among the other cofactors and the prosthetic group biosynthesis pathways, genes encoding folate, pantothenate, coenzyme A, ubiquinone, thiamine, pyridoxine, protoheme have been lost. As a result, Buchnera is dependent on the biosynthesis of some of these latter compounds by the aphid, although folate and nicotinate are present in the aphid diet and probably transported into Buchnera.
Buchnera has lost the genes for gluconeogenesis, the entire enzymatic equipment required to biosynthesize cell surface components (including lipopolysaccharides and phospholipids other than cardiolipin), as well as the regulator genes and genes involved in the defense of the cell.
The symbiotic function of Buchnera should include many compound exchanges, which makes the small number of annotated transporters in Buchnera astonishing. For the transportation of small molecules, Buchnera has retained a few ABC transporter genes, a phosphoenolpyruvate carbohydrate phosphotransferase (PTS) sugar transport system for mannitol and glucose, a porine system, as well as a few non-specific transporters. The conserved flagellum proteins may serve as a transport system for macromolecules in Buchnera (as discussed above).
The Buchnera gene repertoire may have been completed by the transfer of genes into the aphid chromosomes, following a process similar to that observed for the symbiogenetic integration of chloroplasts and mitochondria. However, in contrast to organelles that have evolved from symbiosis with a single-celled ancestor, Buchnera have few contacts with the host germ line, except for brief stages in the life cycle following egg infection. The annotation of the sequenced genome of the pea aphid has made it possible to carry out analyses that confirm the small number of endosymbiont-host gene transfers [75,76].
The sequencing of numerous organisms has made it possible to carry out an automated reconstruction of metabolic networks from genomic data. Hence, the metabolic networks of the four sequenced strains of Buchnera have been inferred and deposited in a dedicated database called SymbioCyc (http://pbil.univ-lyon1.fr/software/symbiocyc/, together with those of 46 other bacteria, including free-living, parasites and endosymbionts).
The evolutionary history of Buchnera can be inferred by comparing the structural properties of its metabolic network to those of E. coli. More precisely, we analyzed the link between the gene repertoire of Buchnera, as a subset of that of E. coli, and its metabolic network structure, in order to identify possible selective constraints linked to the topology of the network.
The metabolic network of BAp was characterized by several graph measures associated with both the reaction and compound graphs (Table 1). The number of connected components2 and the network density3 make it possible to define the connectivity network of Buchnera relative to that of E. coli. The compound graph for Buchnera has fewer connected components (15 vs. 60) and a greater density (0.009 vs. 0.004) than E. coli. The biggest connected components on each graph identify the main structural properties of metabolic graphs, as they correspond to more than 90% of each network (90% of nodes and 92% of edges for Buchnera, and 87% of nodes and 95% of edges for E. coli). The biggest connected component of Buchnera is twice as dense as that of E. coli despite having the same diameter.4 The compound graph of Buchnera seems to be more connected than that of E. coli. The proportion of the shortest paths confirms this: in Buchnera, there are more paths for node pairs as the proportion of shortest paths is lower (42% vs. 55%). Intriguingly, the average path length is greater in Buchnera than in E. coli.
When comparing the reaction graphs, we noticed that the two graphs had the same diameter, but this time the E. coli graph was more connected (density and average number of neighbors). More generally, when comparing the distribution of several topological features in Buchnera and E. coli reaction graphs (Fig. 6), we observed a striking similarity of shape, suggesting that the reaction graph of Buchnera had conserved many of the structural topological properties of its ancestors.

Comparative analysis of the distribution of topological measures for the reaction networks of Buchnera aphidicola Ap and Escherichia coli. The Buchnera and Escherichia coli graphs were obtained from SymBioCyc.
The global parameters of the previous systems analysis (diameter, connectivity, density) give a synthetic overview of the compound and its reaction graphs, and make it possible to carry out comparative genomic analyses in order to study the evolution of the structure of the metabolic networks. It is still very difficult to provide a biological interpretation of these systemic parameters. Global parameters, such as density and diameter, cannot be directly related to the metabolic capabilities of an organism. However, biological interpretations are only possible for local parameters, such as centrality, which is associated with essentiality (e.g., ATP is one of the most central cofactors in metabolic networks). The biological interpretation of systemic parameters requires understanding the link between local properties and the global descriptors of the network, a link that has not yet been established, even in graph theory [77].
4 Evolution of the symbiotic function in Buchnera
The persistence of symbiosis over the course of aphid evolution for more than one hundred million years illustrates the success of this cooperative association. As in all symbioses, cooperation and conflicts have shaped both partners [78]. Aphids feeding on phloem sap are not able to reproduce successfully without their symbiotic bacteria, and they have also developed specialized mouthparts and metabolism that would make it difficult for them to change diet. Buchnera has evolved to use precursor molecules from the host, via the bacteriocytes, and has lost its ability to grow outside aphid cells. The evolution of the symbiotic function of Buchnera has constrained the long-term ecological implications of the diversification and adaptation of the aphids. Hence, loss of the pathway for assimilating inorganic sulfur in BSg (because grass phloem sap is naturally rich in organic sulfur compounds) has forced this aphid lineage to remain on grass throughout its subsequent evolutionary future [7]. In BAp, Thomas et al. [79] have revealed the coupling between the histidine and purine biosynthesis pathways via the partial truncation of the purine biosynthesis pathway prior to AICAR (5-aminoimidazole-4-carboxamide ribonucleoside). The coupling between this essential amino acid biosynthesis pathway (which has been exported to the host) and bacterial growth prevents the bacterium from exploiting the nutrients it obtains from the host without reciprocating.
The isolation of Buchnera within the bacteriocytes precludes the incorporation of any new genetic material from external bacterial diversity, and it is puzzling to observe that Buchnera has lost some metabolic capabilities that could benefit its mutualist host. Are Buchnera symbioses being driven to extinction? Following the process of mutation accumulation and of Muller's ratchet, it seems that aphid endosymbionts may irreversibly erode their genome and their symbiotic function, leading to the extinction of both endosymbionts and aphids [29]. It is still unclear if there is a minimum genome size and gene repertoire required to conserve a functional symbiotic lineage. The genome of Buchnera has not yet dwindled to the 160 kb of the extremely reduced genome of Carsonella rudii [80]. Nevertheless, the selection constraints on genome reduction are related to the symbiotic relationship, and different endosymbionts will follow different evolutionary routes depending on the hosts and ecological niches they occupy.
Gene transfer is quite rare in this model, in which the aphid germ line is not permanently infested by the endosymbiont [75,76], and so Buchnera is not expected to follow the organelle evolutionary route. However, aphid safety may rely on secondary symbionts that could overcome the deficiencies of Buchnera. Such endosymbiotic replacements have been demonstrated in another symbiosis in the Dryophthoridae weevils [81]. The experiments of Koga et al. [25], showing that secondary endosymbionts invade bacteriocytes and can compensate for the loss of Buchnera in A. pisum, as well as the nutritional complementation of BCc by the secondary endosymbiont [82] in C. cedri further support the replacement hypothesis.
Due to the variable demand of the aphid, the symbiotic function of Buchnera must be regulated in some way. However, at present, nothing is known about the signaling of the host demand or the response mechanisms of the endosymbionts. According to the work of Moran et al. most of which was done on BSg [38], specific transcriptional gene regulation is not thought to occur. This work has revealed the absence (or diminution) of transcription regulation in Buchnera, as compared to free-living bacteria placed in comparable situations, when the aphid hosts were stimulated by a heat shock [61] and when aphids were fed with diets in which the amino acid composition had undergone several changes [83]. In BAp, significant (albeit weak) transcriptional regulation mechanisms have been revealed using more physiological nutritional perturbations by rearing aphids on specific artificial diets [55]. Currently studies attempting to decipher the transcriptional response of Buchnera include the separate analysis of the maternal and embryonic bacteriocytes, as these two bacterial populations are known to have differing nutritional requirements [84]. Another approach includes an analysis of the dynamics of the response to a specific nutritional stress (Viñuelas et al., manuscript in preparation). Broad gene expression shifts involving global transcriptional regulators may also be involved in the regulation of the symbiotic function of Buchnera, but no direct evidence for this has yet been provided. Finally, Thomas et al. [79] used FBA modeling to show that any modification of the precursor input (i.e., the ratio between carbon and nitrogen) can have a substantial impact on the output of the essential amino acids produced by Buchnera. This may enable the aphid host to control the production of essential amino acids of Buchnera by restraining the availability of certain precursors within the bacteriocytes and/or across the symbiosome membrane.
Analysis of the metabolic network of the bacterium highlights the evolutionary constraints that have shaped it. Buchnera has conserved the enzyme repertoire required to produce essential amino acids from several precursors that are abundant in the cytoplasm of the bacteriocytes. Some of these precursors are aphid waste products, especially NH3, which is recycled through the symbiotic compartment to synthesize essential amino acids. As Douglas [78] has pointed out, both partners benefit from microbial consumption of animal nitrogenous waste compounds. The aphids benefit, because Buchnera acts as a sink for potentially toxic nitrogenous waste compounds, and the symbiont benefits from access to a source of the nitrogen it requires for growth. The real cost for each of the partners has not been yet evaluated, e.g., the exchange in terms of nitrogen, carbon, oxygen and phosphate flux required for the relative growth and reproduction by the two interacting partners.
Another important aspect of research into the symbiotic function of Buchnera involves elucidating how the metabolic exchanges occur between the partners. As Buchnera transporters appear to be sparse, additional transporters from the host may be involved via the symbiosome membrane, which sometimes fuses with the bacterial membrane at some contact points (Fig. 1). Degradation of the endosymbionts may also be a mechanism used by the host to extract some of the substances produced by Buchnera, as well as to control for the number of endosymbionts. This aspect has not yet been clearly elucidated.
Systemic analyses fed by high-throughput biology now make it possible to reach a better understanding of the symbiotic function of Buchnera. Buchnera does appear to have been shaped by intracellular life, and has adapted to being controlled by the homeostasis of its host. A severely curtailed genome, composed preferentially of multifunctional genes, a fragile, non-redundant metabolic network at the limit of viability for a bacterium [79,87], and the adaptation response to the amino acid needs of the aphid host, all support this conclusion. The recent sequencing of the pea aphid genome [75] now opens the way to extending the analysis of the complete symbiocosm (i.e., the aphid and its procession of symbiotic bacteria), which should allow us to identify genes or key pathways as potential targets for agricultural control of these devastating insects.
Acknowledgements
This work was supported by the “Programme Fédérateur INRA de Biologie Integrative, AgroBI 2006” and by the joint ANR-BBSRC-Sysbio 2007 METNET. The authors of this paper belong to the BSMC group (http://bsmc.insa-lyon.fr). We are grateful to Monika Ghosh for her critical reading and English corrections.
1 Although their lineages are substantially divergent, Buchnera aphidicola is the species designation for the symbionts in all aphids, and this should be abbreviated as B. aphidicola. However, as the genus name “Buchnera” is commonly used to designate this symbiotic bacterium, we use this term rather than “B. aphidicola” in this review.
2 A network is called connected if any two of its vertices are linked by a path (a set of edges leading from one vertex to another). The maximal connected subnetworks of a network are called its components. The number of connected components indicates how strongly the network is connected [77].
| Compound graph | Reaction graph | |||
| E. coli | Buchnera | E. coli | Buchnera | |
| Clustering coefficient | 0.043 | 0.034 | 0.415 | 0.306 |
| Connected components | 60 | 15 | 15 | 8 |
| Diameter | 18 | 18 | 8 | 8 |
| Shortest paths | 466964(55%) | 59388(42%) | 793017(90%) | 86735(90%) |
| Characteristic path length | 5.622 | 7.183 | 2.54 | 2.797 |
| Avg. number of neighbors | 3.417 | 3.014 | 97.084 | 26.168 |
| Number of nodes | 1047 | 416 | 935 | 310 |
| Number of edges | 2285 | 758 | 56698 | 4975 |
| Density | 0.004 | 0.009 | 0.13 | 0.104 |
| Isolated nodes | 4 | 1 | 13 | 7 |
3 The network density or the edge density is the proportion of potential edges that the network actually has [77].
4 The distance between two vertices in the network is the length (the number of edges) of the shortest path between them. The greatest distance between any two vertices in the network is the diameter of the network [77].


