1 Introduction: Living organisms, complex systems and systems biology
During the last decades an impressive amount of information has accumulated regarding the molecular mechanisms at the basis of both plant and animal development. Although we have gained significant new insight in the nature of the genetic networks and cellular processes at work, major questions have remained unsolved. It has been difficult, for instance, to define the precise role of genetic determinants in controlling morphogenesis and the control of shape is currently a major and largely unresolved issue in both animal and plant biology. High throughput approaches have helped to determine the expression patterns of thousands of genes during development, but the results are difficult to integrate and sometimes impossible to interpret fully. We might understand the basics of cell division and growth, but how to understand the collective behaviour of thousands of cells in a tissue? In this context the concept of complex systems has been introduced in a discipline called “systems biology”. Living organisms, as complex systems, can formally be defined as sets of entities that interact according to simple local rules and whose collective activity is non-linear. These interactions lead to the emergence of new properties at a higher level of organization that cannot be deduced from simply adding up local behaviour. Thus molecules interact and assemble into cells with particular properties, which in turn generate interaction networks to form tissues and organs. Because studying living organisms occurs at such a high level of integration, the developmental biologist usually does not have access to the rules that drive the initial interactions. Systems biology offers a framework to test such rules and check whether they are able to generate the expected ontogeny. There is often feedback between levels of organization. For example, the interactions between cells can generate gradients of signalling molecules that span whole tissues. This overall distribution of molecules at the level of a whole tissue, will in turn feed back locally on the behaviour of individual cells, reacting to specific concentrations. Systems biology offers a framework to analyse such feedbacks. More generally systems biology analyses processes like plant development at different scales, describing not only the properties of individual cells but also their interactions. The complexity of the information involved is such, that it cannot be understood and integrated on a purely intuitive basis. For this reason, modelling approaches have become more and more important to study and analyse biological systems. In this review we will illustrate how these approaches can help to understand complex problems such as plant morphogenesis. Because of space constraints, it will not be possible to discuss all aspects of plant development within one review. We will, therefore, limit ourselves to morphogenesis at the shoot apex. More in particular we will focus on meristem function.
2 Morphogenesis at the shoot apex: A central role for the meristem
2.1 The plant meristem as a complex system
Plant meristems are populations of undifferentiated stem-cells that continuously proliferate both to generate tissues and organs. Since meristems can modulate their activity in response to external cues, they provide the developmental flexibility that allows plants to modulate their shape and development in reaction to prevailing conditions (see reviews: [1–4]).
A typical plant contains distinct meristems. Apical meristems, positioned at the tip of the shoots and roots, initiate aerial and underground organs respectively. Along the stems and roots more diffuse secondary meristems exist which are responsible for secondary thickening of these structures.
As mentioned above, we will focus here on the morphogenetic processes that lead to the formation of aerial organs, which is due to the activity of shoot apical (SAM) and floral meristems. Starting from a brief overview of our current knowledge on meristem structure and molecular regulatory networks, we will subsequently discuss how modelling approaches have helped to better understand morphogenesis at the SAM.
2.2 The shoot apical meristem: A dynamic structure with a stable organization
All SAMs have a number of basic structural characteristics in common, which can be identified by simple histological observation (e.g. [1,2]). One of the most prominent features perhaps is the presence of one or several distinct cell layers – called the tunica – which cover the internal tissues – called corpus – of the angiosperm meristem. The tunica layers are kept separate as their cells most frequently divide in anticlinal orientations causing daughter cells to remain in the same layer as their parent. If present, the internal tunica layer – called L2 – usually disappears during organ initiation, when L2 cells start to divide in random directions as well.
Superimposed to this organization into tunica and corpus is a partitioning into zones. These zones, characterized by subtle differences in cell characteristics have particular functions. At the meristem summit there is a small group of slowly dividing cells, called the central zone, which have a stem cell function and ensure meristem maintenance. The growth rates at the meristem summit usually differ considerably from those at the periphery where cells accelerate their proliferation rates. It is in this zone that the new lateral organs are initiated. Primordium initiation involves the definition of a group of founder cells, which will subsequently form the lateral organ. Both clonal analysis and direct in vivo studies carried out on the model species Arabidopsis have shown that the epidermal layer of the flower primordia can be traced back to small groups of about four to ten cells in the L1 at the periphery of the meristem summit, probably just outside the central zone [5,6]. Once the founder cells have been defined, cell proliferation and growth rates increase further resulting in the formation of a primordium that rapidly grows out. It is limited by the organ boundary region, where cell expansion is reduced.
3 Analyzing shoot apical meristem function using modelling approaches
In view of its importance, the SAM has been extensively studied. Genetic analysis has identified the major molecular determinants of meristem function, including transcriptional regulators and signalling cascades involved in meristem maintenance and organ initiation. Advanced live imaging techniques have started to give a more and more detailed view of the dynamics of growth and gene expression. In the following paragraphs, we will discuss three examples where modelling approaches have helped to analyse these data.
3.1 Modelling gene regulatory networks
Extensive genetic screens have identified a series of transcriptional regulators involved in meristem function, organ initiation and outgrowth. Without giving an exhaustive survey of the regulatory network, we will simply provide a brief description of the main factors implicated to illustrate the complexity involved (for reviews: [2–4,7]).
At the heart of preserving the shoot meristem is the transcription factor WUSCHEL (WUS) which is required to maintain the meristem as in corresponding mutants a SAM is initiated but arrests after having produced a few organs [8,9]. Its precise targets and regulators are largely unknown, although it has been shown to promote the expression of the signalling peptide CLAVATA3 in a non-cell-autonomous way and repress several cytokinin response factors belonging to the type-A ARABIDOPSIS RESPONSE REGULATOR family ([10,11], see below).
At the periphery of the meristem the initial recruitment of the organ founder cells appears to be the result of two antagonistic processes. First, the homeodomain protein SHOOTMERISTEMLESS (STM), in combination with several members of the so-called CUP SHAPED COTYLEDON (CUC)-family of transcription factors define meristematic identity, preventing cells from being recruited by the young organs [12–17]. Therefore, as a new organ is initiated, these meristematic identity genes are switched off. While the initial repressors of these genes in the incipient organ are still unknown, several regulators including ASYMMETRIC LEAVES1 (AS1) maintain the repressed state of meristem identity factors such as STM in the growing primordium [18]. The meristem can either generate leaves and lateral meristems or it can initiate flowers. The identity of the lateral organs produced depends on the activity of LEAFY (LFY), which, by interacting with different transcription factors involved in meristem function such as WUS or AGAMOUS (AG), plays a major role in plant and flower architecture [19,20].
The patterning of the flower itself also largely depends on transcriptional regulation (for reviews, see [21,22]). This includes genes that control organ identity, organ number, organ boundaries, local patterning (e.g. polarity) and local control of cell division. Of particular relevance are the organ identity genes, APETALA1 (AP1), CAULIFLOWER (CAL), PISTILLATA (PI) SEPALLATA1 (SEP1), and APETALA3 (AP3), which direct the development of sepals and petals [21,23,24]. Other relevant genes are upstream regulators of the organ identity genes (such as UNUSUAL FLORAL ORGAN (UFO) and LFY; [25–28], genes that control the number of sepals and petals formed in each flower (PERIANTHIA (PAN), WIGGUM (WIG) [29–31]), genes required to establish organ boundaries (CUC genes; [14–17]), and genes that participate in organ patterning (KANADIs, YABBYs and the PHB/PHV/REV group; [22,32–36]).
Once the founder cell populations of the organs have been identified, they grow out. Our knowledge on the genetic regulation of this process is more limited. Two cellular processes contribute to the control of final organ size: cell division and cell expansion. However, the various signals that integrate and coordinate these processes in the context of a given organ in terms of growth remain largely unknown. Certain of the connections between the basic cellular components and regulatory networks have been identified. AINTEGUMENTA (ANT), for instance, is strongly expressed in the rapidly outgrowing organ and seems to act as least partially via CYCLIN D, a gene involved in cell proliferation [37]. For petals, several genes were shown to control growth by affecting cell proliferation and/or cell expansion, some in an organ specific manner [38]. Some of these genes (i.e. JAGGED, ANT, ARGOS) were shown to affect petal growth by positively regulating cell proliferation [37,39,40], whereas other genes (BIGBROTHER, KLUH and DA1) were shown to control final organ size by negatively regulating the duration/period of cell proliferation [38,41,42]. BIGPETALp, which is regulated downstream of the flower organ identity genes, was shown to limit petal growth by controlling cell expansion [43].
In addition to transcriptional control, post-transcriptional regulation by microRNAs (miRNAs) plays an important role in meristem function. These include, for example, the miR164 and miR156/157 families, which respectively target the CUC and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes (e.g. [44,45]).
The very extensive molecular genetic analyses allow us to draw a model of the regulatory network controlling meristem function (Fig. 1). While this model provides a summary of our current knowledge, it also points at major shortcomings in the available data. First, the network is unfinished. Indeed, although many of the major players have undoubtedly been identified, the picture is not complete as illustrated by recent transcriptomic analyses of different domains of the SAM, identifying new markers for the central stem cell populations and revealing novel expression sub-domains [46]. In addition, the nature of the interactions presented in the model is very diverse as their relationships, in many cases deduced from genetic interactions, are often indirect and poorly characterized. Third, very little quantitative data are available and often nothing is known on protein or RNA concentrations, stability, interactions between molecules, reaction rates, etc. As a result it becomes very difficult to fully understand how the network operates or even formalize our knowledge in the form of a regulatory network model. Nevertheless several attempts have been made to produce such models. Since quantitative details on the molecular reactions are not available, these models have been restricted to so-called Boolean approaches, where the nodes of the network (genes, proteins, metabolic activity, etc.) are simply on or off (for review: [47]). Gene interactions, for example, are represented as simple positive or negative effects, either promoting or inhibiting the activity of targets. This might seem simple but even this approach already poses a number of problems. For instance, when a particular gene receives both negative and positive inputs, how to decide which of the two will prevail? Therefore, usually some level of quantified information (e.g. indicating that one input is stronger than the other one) is integrated to these models. Since the strength of a particular interaction is often unknown (and even indirect) this has to be estimated, often from indirect evidence such as double mutant phenotypes analysis. Once the network model has been constructed, it can be used to determine how coherent and complete the available information is. Thus the model should be able to reproduce so-called stable states where particular combinations of genes are switched on or off, corresponding to the gene activities in particular cell types. Boolean models give a very raw representation of reality, remain often very descriptive and have limited predictive power. They are, however, often the only way to represent our current knowledge in a formal manner and do allow us to identify potential contradictions or missing information. This type of models has notably been used to analyze cell cycle networks (e.g. [48]), and flower development [49,50].

Part of the regulatory network controlling meristem function. The figure summarizes part of the available knowledge on the interactions between a set of meristem regulators (in circles). The links between the different factors are represented as inhibitory or stimulating (arrows), for details see text. This figure represents a considerable amount of information, yet represents a number of shortcomings. Arrows, for instance represent different types of interactions (from direct transcriptional regulation to indirect non-defined relations and hormone transport), no quantitative or spatial information is represented. Gene names: WUSCHEL (WUS), CLAVATA (CLV), SHOOTMERISTEMLESS (STM), CUP SHAPED COTYLEDON (CUC), ASYMMETRIC LEAVES1 (AS1), LEAFY (LFY), AGAMOUS (AG), APETALA1 (AP1), SEPALLATA (SEP), APETALA3 (AP3), PERIANTHIA (PAN), AINTEGUMENTA (ANT), MONOPTEROS (MP), PIN-FORMED (PIN), TERMINAL FLOWER (TFL). Hormones: Giberellins (GA), Cytokinins, Auxin. See Sablowski, [4]; Rast and Simon, [7].
In the flower, the interactions (activation and repression) between fifteen floral mastergenes build a network the topology of which could generate four attractors (i.e. a convergence to four gene activity states independent on the initial configurations), associated to the four floral organ identities, as predicted by the ABC model. The validity of the model was further confirmed by the observation that it could also reproduce the mutant phenotypes [49]. Interestingly, minor alterations to the interaction rules did not significantly alter the outcome of the simulations, suggesting that the robustness of flower ontogeny among various species resides in the structure of this network [49].
3.2 Modelling cell–cell interactions
Two signalling networks have been extensively studied during the last years. The first involves the regulation of meristem maintenance, the second the hormonal regulation of organ initiation. Modelling has again played an important role in further analyzing these networks. Compared to the Boolean networks mentioned above, the simulation of these interaction networks represents an additional difficulty as it also has to take into account spatial aspects. In addition, since signal gradients are often implicated, a simple “on” or “off” status of the signal is not a realistic option. Therefore, differential equations are used to calculate the changes in signal (e.g. [51,52]).
The first example concerns meristem maintenance which, as mentioned earlier, depends on the homeodomain transcription factor WUS. WUS itself interacts with a small signalling network involving the CLAVATA (CLV) ligand/receptor complex [22]. The small WUS expressing domain, a small subset of cells inside the meristem, is surrounded by a population of cells expressing the receptor kinase CLV1, which in its active form, restricts WUS expression and the size of the WUS domain. WUS itself activates a small peptide, CLV3, in the cells just above its expression domain. CLV3 probably diffuses in the intercellular space to the CLV1 domain, thus activating the receptor kinase. Thus, when WUS concentrations rise, CLV will become more active and tend to reduce stem cell identity. Conversely, a strong inhibition of WUS will automatically lead to a reduction in CLV3 signalling, thus weakening the repressive action [10,22,53,54]. Several modelling studies have investigated how this negative-feedback loop not only permits meristem maintenance, but also limits important changes in meristem size (e.g. [55,56]). Although it is one of the best analysed regulatory units in the meristem, experimental data for this network is still missing. Notably, the mechanism behind the spatial locations and dynamics of these expression domains is subject to interpretation. Jönsson et al. [55] therefore designed predictive mathematical models as useful tools for determining which hypotheses best explain the observed gene expression dynamics. These models had the form of two dimensional longitudinal or transverse sections through the meristem. One of the models proposed, used a reaction–diffusion mechanism, in which an activator induces WUSCHEL expression. This model was able to organize the WUSCHEL expression domain. In addition, the model predicted the dynamical reorganization seen in experiments where cells, including the WUSCHEL domain, are ablated, and also predicted the spatial expansion of the WUSCHEL domain resulting from removal of the CLAVATA3 signal [55]. Although the presence of such an activator remains to be established, this type of model is a good example of the next generation, which, in contrast to classical Boolean approaches, uses and produces quantitative and spatial information.
A second example where modelling has been instrumental comes from work on organ positioning or phyllotaxis. Several signalling molecules including plant hormones are essential for this process. In particular the phytohormone auxin is a major player. A widely accepted hypothesis proposes that local auxin accumulation at specific locations leads to the initiation of new organ primordia [57–63]. Auxin is actively transported throughout the plant by membrane associated membrane associated transporters of the so-called PIN family which are often localized on one side of the cell. Since neighbouring cells often show coherent PIN localizations, it was proposed that these proteins create fluxes through the tissues, leading to auxin minima and maxima (e.g. [57,64]). Recent observations suggest that auxin maxima created by these proteins are at the basis of organ initiation. Accordingly, genes involved in auxin transport from cell to cell are essential for organ initiation and their mutation severely compromises or even completely disrupts primordium formation (e.g. [57]). This hypothesis was initially based on the visual, qualitative characterization of the complex distribution patterns of the PIN1 protein in Arabidopsis. To take these analyses further, the properties of the patterns were investigated using computational modelling, where real images of PIN distribution were interpreted in the form of cell–cell connection maps in which the putative fluxes of auxin could be analysed [59]. The simulations confirmed that auxin fluxes were likely to be directed to the young primordia, but also revealed previously undescribed properties of PIN1 distribution. In particular, the simulations suggested an accumulation of (inactive) auxin at the meristem summit. This prediction was validated by monitoring the expression pattern of the auxin activity sensor DR5::GFP and by immunolocalizing the auxin in the meristem [59]. Altogether both the experimental and simulation data suggest an important role for this local accumulation of auxin in meristem functions.
In parallel to this analysis of the observed patterns, more hypothetical models tried to explain the basic principles of these apparently very complex patterns. Interestingly these models showed that very simple cellular behaviours can potentially explain the phyllotactic patterns of cell interactions. Both Jönsson et al. [60] and Smith et al. [61] showed that a scenario where cells would simply probe their direct environment and transport auxin to the neighbour with the highest hormone concentration would be sufficient to create the patterns of auxin maxima observed in the real plant (see also [65]). This perfectly illustrates an example of a complex system, where very simple local interactions (cells transporting against a gradient) can lead to patterns (in this case localized auxin maxima at precise positions) at the level of a whole tissue. Conversely, the modelling approach was able to demonstrate how such global patterns can feed back on local (cellular) responses [59–61]. More generally, it also illustrates how useful modelling can be in these cases, as hormone fluxes in a population of hundreds of cells are impossible to predict on a purely intuitive basis.
Another point concerns the validity of the models. Although they reproduced a number of observations such as PIN localization or organ position with accuracy, this is not a mathematical proof they are true. Indeed, more recently Stoma et al. [63] produced an alternative model, based on cells sensing fluxes of auxin instead of concentrations, that also reproduced PIN localization patterns with even greater precision. Models are, therefore, no final proof of concept but rather tools that allow us to test the plausibility of complex hypotheses and to make predictions to be tested in future experiments.
3.3 Towards an understanding of morphogenesis: From genes to shape
In the previous paragraphs we have discussed the genetic regulatory networks and shown how cell–cell interactions could lead to organ initiation at precise positions. The next important question is how patterns of gene expression and signalling molecules are linked to the actual growth patterns we observe.
It is commonly accepted that genetic regulation is at the basis of morphogenesis, but in many cases, the effects of genes on shape have only been defined in a very general manner. Indeed, genes involved in flower formation, for example, have been identified, but at this stage we only know that particular genes (e.g. AP2) are responsible for the formation of a particular organ (e.g. a petal). In addition to the analysis of mutants, inducible expression strategies have been developed to either promote or inactivate certain genes at a particular stage and thus refine the exact contribution of these genes in a given morphogenetic process (e.g. [66,67]). Nevertheless, we do not know, in any detail how the genes influence the dynamic growth patterns leading to organs with particular shapes. This is because mainly qualitative information is available regarding gene function. Indeed, considering growth patterns as outputs of gene action requires detailed knowledge of geometry over time and a quantitative framework for describing geometrical changes [68,69]. A precise framework was formulated by Coen et al. [70] who proposed four types of regional parameters to describe growth and shape:
- • Growth rate, the rate at which a region changes in size (e.g. area or volume);
- • Anisotropy, the degree to which growth occurs preferentially in one direction;
- • Direction, angles at which the directions of growth are oriented. One of these will be the main direction of growth. This type of parameter is obviously related tot the previous one;
- • Rotation rate, the angle through which the region turns relative to other regions per unit time. This is not a local growth property, because there is no change in shape or size.
A precise quantification of these four parameters can be used to accurately describe any changes in shape of any growing systems. How can we express gene activities in these precise terms? Several options are currently being investigated. First, growth patterns can be captured in distinct domains where specific gene combinations are active (e.g. the central zone or organ boundaries). Second, growth patterns can be analysed in mutant backgrounds. Determining when and where particular parameters are perturbed should then provide information on gene function. To measure these variables several approaches have been developed. Coen and co-workers induced coloured sectors induced by a transposon that is activated after a temperature shock in developing petals of Anthirrhinum [71]. Since the temperature shock was given at a particular time, the size and shape of the resulting sectors (corresponding to the descendants of single cells) gave information on both the direction and rate of growth that occurred. In parallel, several groups have developed methods to analyse growth with cellular resolution. Kwiatkowska and colleagues (e.g. [72]) for example have used replicas from the surface of living meristems to analyse the growth patterns in both wild type and mutant plants of different species. More recently, confocal life-imaging has been used to study the dynamics of meristematic cells as well [5,6,58,73]. The advantage of confocal microscopy lies in the possibility to obtain information on the growth patterns of internal tissues as well. The problem here is to provide high resolution 3D images of growing meristems, and to follow the growth patterns of single cells over several days. These techniques are only just beginning to be developed (Fig. 2).
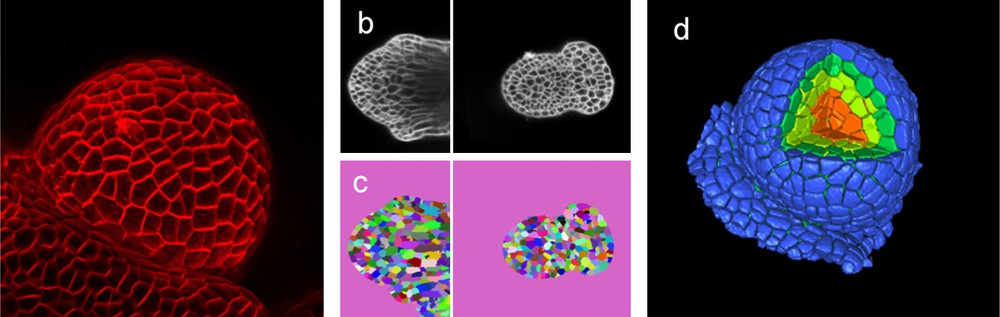
Quantifying growth with cellular resolution. Imaging is at the basis of growth measurement. In this example, living flower buds are visualized in a confocal microscope (a, b). Pictures are then used to identify the individual cells (c) as individual (colour coded) object. Once the pictures have been segmented, a reconstruction can be made (d) where every single cell is identified as a single object that can be measured and analysed. In (d) a section of the reconstructed meristem has been removed to show the internal tissues as well (Das, Fernandez, Traas, Godin & Malandain, unpublished).
Once the general growth patterns have been established, they have to be interpreted and linked to gene function. To address this issue, models are very useful. Indeed, since growth is a complex spatial–temporal process, it is impossible to propose a precise role for a particular gene without simulations to explore the parameter values. Such models not only have to take into account the anisotropy and growth rates of individual cells, but also physical interactions between cells ([70], Chopard et al., unpublished data). Certain aspects of these mechanical models will be discussed in the following paragraph.
3.4 Beyond geometry: modelling physical properties
Once a quantitative description of gene activity in terms of growth is established, the next step is towards a mechanistic approach, where the determinants of the regional and cellular growth parameters have to be studied.
From a mechanistic point of view, the gene regulatory network must somehow interfere with the physical properties of the cells to generate specific shapes of organs. In this context interactions with cellular structural elements such as the cytoskeleton and cellulosic cell wall are central (review: [2], see also: [74]). During growth, cellulose is deposited outside the plasma membrane, thus preventing the wall from becoming thinner. Because of their high tensile strength, cellulose microfibrils are the main determinants of cell wall rigidity. The fibrils are often embedded in a matrix containing other polysaccharides such as hemicelluloses and pectins which binds them together. Therefore, cell wall expansion in the growing cell not only depends on cellulose synthesis, but also on the synthesis and remodelling of the matrix. Thus, work aimed at analyzing the cell wall structure at the meristem has indicated that important modifications in pectin composition are essential for organ initiation [75], regulating the differences in growth rate. In addition to its central role in growth rate control, the cell wall is also the main determinant of cell growth anisotropy. This is because cellulose fibrils are often deposited in highly ordered arrays, preventing expansion in one direction. Because this direction is often parallel to the cortical microtubules of the plant cell, it is thought that the microtubules serve as tracks along which the cellulose synthase complexes travel (e.g. [76]). This raises the question what orients the microtubules. This issue was recently addressed by Hamant et al. [74]. The authors observed that microtubules are often parallel to the predicted stress patterns at the meristem surface. Modifying the forces by changing the shape of the entire meristem or by locally perturbing force patterns using ablations confirmed this observation. This led to the idea that microtubules, by orienting microfibrils parallel to the main stress patterns, would make the cells resist these forces. This, in turn, would be important for certain morphogenetic events, such as tissue folding in organ boundaries or the formation of a cylindrical stem ([74] see also [77,78]). To test the plausibility of this hypothesis, models able to express mechanical properties were designed. To do so, two approaches were considered. The first one consisted of representing the mechanical cell interactions using a mass-spring particle system (walls and cell vertices represented as springs and particles respectively, see Fig. 3). Using this method, the surface of the meristem was represented as a network of springs. To model the feedback of force fields on microtubule orientations, the cells in the model were instructed to stiffen the springs in function of their angle to the main forces. This assumption was enough to reproduce at least qualitatively the behaviour of the microtubules in the model. In addition, the simulation reproduced simple morphogenetic events such as tissue folding at the boundaries of outgrowing organs and the formation of a cylindrical stem [74].

Three-dimensional (3D) models that also incorporate mechanics are important to simulate growth dynamics in tissues. A relatively simple mechanical representation of the tissue represents cell walls as springs. Every spring has a restlength (lo). The real length changes to ‘l’ when the network of springs is put under tension. Growth can be simulated by changing the restlength. This approach can be used to transform an original picture of the surface of a meristem into a model (b). The spring model is mainly a 2D surface in 3D space. More realistic cellular models can be obtained using finite element modelling (c). The small white lines indicate stress patterns (from Hamant et al. [74]).
The mass-spring models were extremely useful in testing the proposed hypotheses. They do have, however, certain limitations. In particular, they only allow the modelling of 2D objects (like the meristem surface) and are less well suited to simulate the dynamics of a full 3D structure. To circumvent these shortcomings, the mechanical properties of the model can be represented at every point of the object by a mechanical tensor to apply the equations of mechanics at each point in 4D. This approach has been implemented efficiently using the finite element method, either on a continuous medium or on a discretized lattice representing the organ/meristem cell walls ([74], Fig. 3). This approach (because it is very demanding in terms of computation) has still to be adapted to calculate the dynamics of a complete 3D virtual meristem, but very efficiently predicted the stress patterns after cell ablation in a virtual tissue composed of one or two cell layers.
Overall, the work, involving simulations, has lead to a picture where microtubule based cell anisotropy controls specific morphogenetic events such as tissue folding or the formation of smooth surfaces and cylindrical stems. Interestingly, this can be uncoupled from the overall control of growth rate. When microtubules are removed using drugs, growth patterns are maintained and cells continue to differentiate. This suggests that the auxin-based induction of organs operates, at least partially, independently from the control of anisotropy [74]. To come back to the previous paragraph where we discussed the genetic control of geometry, the question becomes how genes control microtubule orientations and auxin-based cell wall expansion (Fig. 4).

From gene to shape. A major question is how genes control the shape of organs like the young growing flower presented here. Changes in shape can be described and quantified using two major parameters, growth rate and anisotropy. These parameters allow us to express gene activity in terms of geometrical values. The two parameters in turn depend on cell wall synthesis and microtubule orientation respectively.
4 Conclusion and perspectives: Towards a multiscale model of morphogenesis
The complex datasets generated in multiple experiments, combining dynamic expression patterns with dynamic changes in cell shapes, are impossible to analyse using simple visual inspection and intuitive interpretations. Therefore, to integrate the data and to formulate complex hypotheses in silico modelling tools are becoming increasingly important (see also: [79,80]). Several advances in addressing this issue have been made. Gene regulatory network models for developmental modules in a multicellular environment have been used to investigate molecular mechanisms for meristem maintenance, transport models for auxin have been applied for modelling phyllotaxis, and mechanical models have been used to represent physical properties. In several cases, confocal microscopy data have been directly integrated with the models and model parameters have been directly extracted from experimental data.
The creation and further development of these models is a challenging task and there are several important problems that need to be addressed in our modelling approaches.
First, the amount of expression pattern data generated will lead to large models of gene regulatory networks. Beyond the classical approach at a single cell level, these models will have to explain the spatial organization of gene expression in the tissue. This requires the development of a new modelling framework that reflects the two-level structure of the gene interaction network, i.e. single-cell networks integrated within a network of interacting cells at tissue level. This novel framework will consist in a trade-off between existing formalisms, such as discrete models, mostly used at the single cell level, or the partial differential equations models usually used to model reaction diffusion in continuous spatial media. These two extreme cases will have to be tested, as well as different choices of intermediate, hybrid systems involving both discrete and continuous terms, such as the well studied piecewise-linear models [81,82].
Second, as shape is affected by gene expression patterns and vice versa, a virtual tissue should couple the genetic regulation of cell activities with a physical model of the cell that is capable of growth and division. Such a model will make it possible to derive tissue deformation as a consequence of the mechanical constraints due to differential cellular growth triggered by adjacent domains with different gene expression.
To check the model outputs detailed information will be required on a range of parameters such as gene product levels, expression patterns and growth dynamics. Therefore, in parallel to these modelling efforts, work carried out at the cellular and subcellular level should become increasingly quantitative. Determining the cellular concentrations of hundreds of proteins, measuring the growth in tissues with cellular resolution will on its own be a daunting task.
All these issues are challenging but we believe that they are essential to decipher plant development and the complex feedback loop between gene and shape, mediated by physical forces and biochemical signals. Mechanistic models with cellular and molecular resolution should be an extremely useful complement to models currently developed for whole plants and which are often still very descriptive. The ultimate goal will be to create a multiscale, mechanistic framework that should allow us to develop powerful predictive models of plant development.


