1 Introduction
Two vital functions are devoted to roots: anchoring plants into the soil, and providing them with water and inorganic nutrients. Since water and nutrients are often not homogeneously distributed in soil, plants must optimize the soil exploration with their root system. A striking example of such an optimization is illustrated by Drew's experiments (1975) on barley, showing that lateral root development is locally stimulated by a higher supply of nitrate or phosphate [1]. Other environmental clues condition the direction and the extent of root growth: light, gravity, water gradient, pathogens…
Recent publications have pointed to the role of the root cap located at the extremity of the root tip (for reviews, see [2,3]), which appears to be a key component in the perception and/or integration mechanisms related to some of these regulations. It must be noted that Charles Darwin and his son Francis pinpointed the functional importance of the root apex already two centuries ago. They emphasized in their book “The power of the Movement of Plants” published in 1880: “It is hardly an exaggeration to say that the tip of the radicle [...] acts like the brain [...]”. At that time, their work was centred on the study of the perception of the gravity, but it also identified the root tip as an area involved in hydrotropism and thigmotropism.
This review article intends to summarize the knowledge related to the root cap structure and development, by focusing mainly on the plant model Arabidopsis. We have highlighted recent results revealing its crucial role in root growth, but also as a major sensing [4] site for several environmental clues.
2 Root cap formation and maintenance
2.1 Basic root organization
Root cross sections show that this organ is composed of five major tissues. These are organized following a radial symmetry, and each tissue contains one or several cell layers (depending on the species). Starting from the centre of the root, we find: the stele (or central cylinder), the pericycle, the endodermis, the cortex and the epidermis (Fig. 1). These tissues are further divided into distinct cell types. For instance, two types of vascular tissues (xylem and phloem) are found inside the stele. They are organized in the form of alternating poles: two to four poles per tissue in gymnosperms and dicotyledons (diarch, triarch or tetrarch arrangement) or even more in the monocotyledons (polyarch arrangement) [4]. In Arabidopsis thaliana, tissues are organized in single cell layers except for the cortex (which generates a second layer three weeks after germination) and central cylinder (which has a diarch pattern) [5,6]. This simple radial organization established during embryogenesis is conserved all along the root growth thanks to the root apical meristem (RAM) activity [7–9].
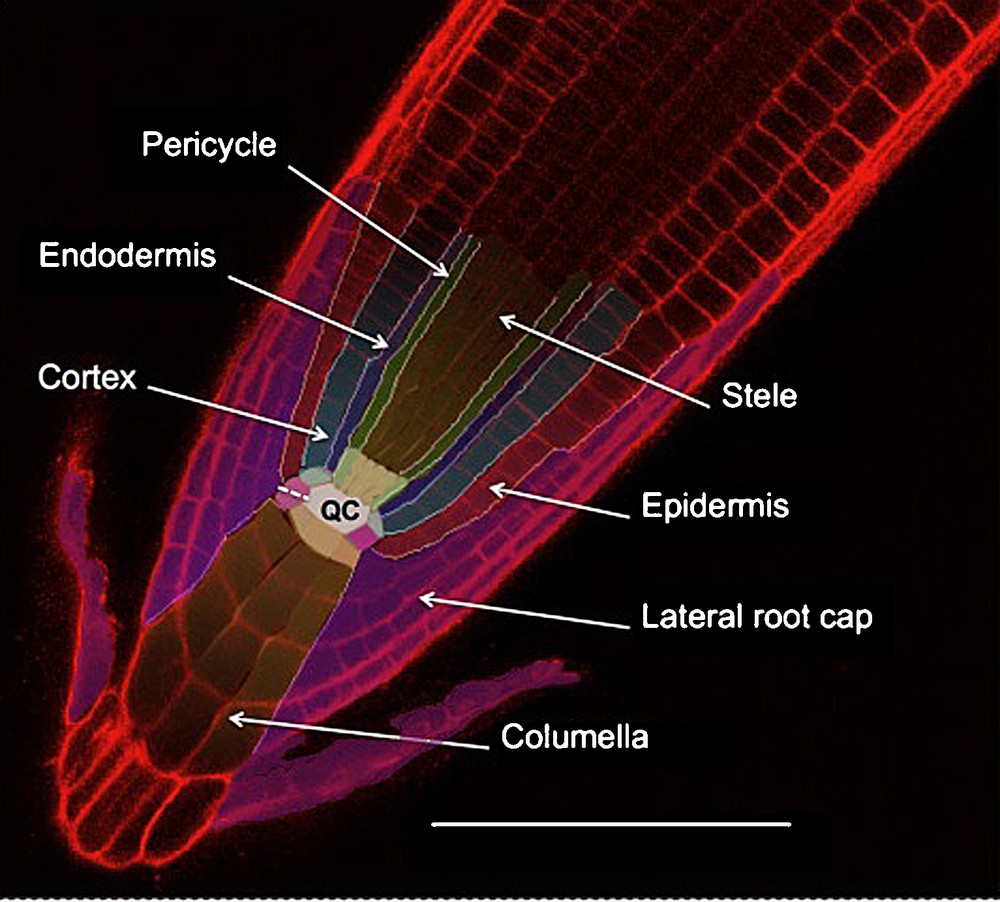
Arabidopsis root tip structure; confocal imaging of root stained with propidium iodide. Initial cells are colorized slightly paler than their corresponding tissue. The quiescent center (QC) is in white. The dotted line indicates the lateral root cap/columella initial anticline division. The scale bar is 100 μm.
The RAM is constituted by two groups of undifferentiated cells: the quiescent centre (QC) and the initials that surround the QC. They can be distinguished by their mitotic activity: high for the initials and very low for the QC [5,10]. Each of the different cell files that constitute the root tissues can be traced back to a single initial in the case of “closed meristems” (such as the ones from Arabidopsis thaliana) (Fig. 1) or to a group of initials in the case of “open meristems” (such as in Pisum sativum) [5,11,12]. Clonal analysis of cell fates in the root tip shows indeed that each initial generates one or two cell files [7,8,13] giving rise to distinct tissues (Fig. 1) [5,7,14–16]. The number of QC and initial cells varies considerably depending on the species: for example, there are four QC cells in Arabidopsis, but 1000–1500 in maize. A highly conserved pattern of division and arrangement around the QC is responsible for maintaining the post-germination root radial organization [5,14–16].
2.2 Root cap structure
In the majority of spermatophytes and pteridophytes, the root tip is surrounded by yet another tissue: the root cap (for a review, see [3]). The root cap forms a little dome of cells covering the RAM. This tissue is strictly organised into two parts: the columella (in central position), and the lateral root cap (LRC) around the columella (Fig. 1). Histological observations show that the columella itself is composed of two different cell types: the statocytes, which are mostly located in the internal layers of columella (and occasionally in some lateral root cap cells), and the secretory cells constituting the LRC and the external columella layer [5,12,18].
The root cap releases secretory cells continuously during the root growth [19–22]. Similarly to other root tissues, root cap cells are generated by the RAM [12,14,15]. Interestingly, the variation of RAM activity occurring during plant development explains the variations observed in root cap sizes [23] corresponding to root diameter variations. Nevertheless, these variations occur within limits that are strictly dependent on the plant species. For example, Arabidopsis thaliana root cap contains 180 to 260 cells [5,14,25] while in Pisum sativum, root cap is formed by 4000–21,000 cells [23].
2.3 Embryonic origin of the root cap
Clonal and histological analyses have shown that root cap precursor cells can be traced back to the embryonic early heart stage [13]. After fertilization, the first division of the zygote is asymmetric and gives rise to the terminal cell (the mother cell of the embryo) and the basal cell (the mother cell of the suspensor, a structure dedicated to the nutrition of the embryo).
At the “early heart” stage, the hypophyse (the suspensor cell connecting the embryo with the rest of the suspensor) divides into:
- • the precursor cell of the QC;
- • the first root cap cells (Fig. 2).
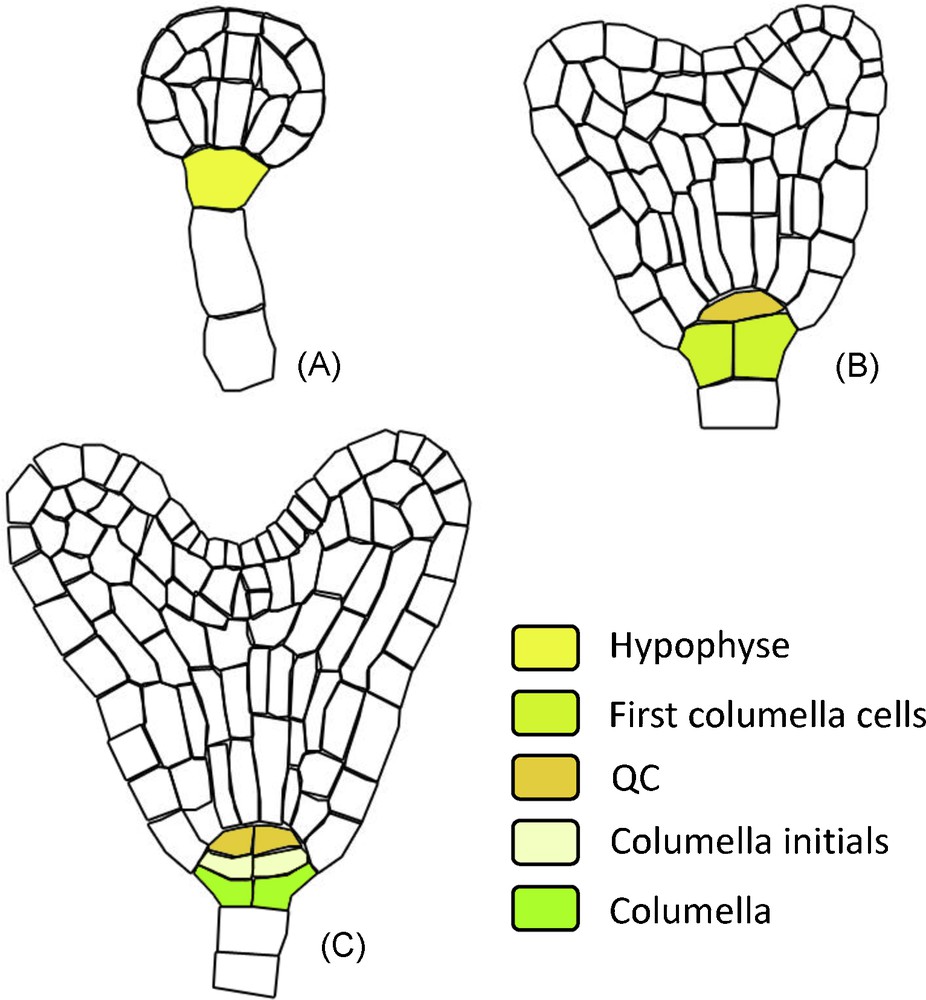
RAM ontogeny: structure derivated from the hypohyse. During embryo development the successive hypophyse divisions gives rise to the QC, columella initials and columella cells. The different developmental stages are annotated: A. Globular stage. B. Early heart stage. C. Mid heart stage.
After this stage, the first columella initials divide to produce additional columella layers inside the embryo [13].
In the mature embryo, the columella cells contain amyloplasts. Interestingly, these organelles at this stage do not sediment according to the gravity vector (contrary to what is observed later on in the case of statocytes), and they are randomly distributed inside the cells. During the dormancy, the amyloplasts starch is degraded, and these amyloplasts display a proplaste-like structure. After only two hours of seed imbibition, all the columella cells differentiated into statocytes following the same process as described below [26].
2.4 Root cap maintenance after embryogenesis
After germination, the columella and the LRC cells are continuously renewed by RAM activity thanks to two different sets of initials. The columella initials constitute a mono-layered cell disc located between the QC and the collumella. Their asymmetric anticlinal divisions generate the columella cells [5,14,15].
The lateral root cap initials form a mono-layered ring surrounding the RAM [5,6]. In the case of closed meristems, the LRC and the epidermis are produced by the same initials after an asymmetric periclinal division followed by anticlinal divisions [14,15]. The analysis of 27 species with closed root meristems distinguishes three types of spatio-temporal division patterns producing: spiralling cones (Arabidopsis thaliana), overlapping arcs (Gossipium hirsutum) or cylinder cones (Linum grandifolium) [14]. In species with open root meristems, the epidermis and the LRC are produced by different sets of initials through anticlinal divisions [27].
Just after the division of columella initials, the proplasts in very young columella cells start to accumulate starch inside amyloplasts, which increases their weight, and, in conjunction with the decrease of cytoplasm density, it induces their sedimentation inside the statocytes [18,23,29]. Then, the starch content of amyloplasts degradates with the progressive differentiation of statocytes into secretory cells, and numerous small vacuoles aggregate to generate a single compartment. In these cells, the number of organelles increases, particularly golgi apparatus and vesicles involved in secretory functions. In LRC, cells only differentiate into secretory cells. When fully differentiated, the root cap secretory cells increased their initial volume by a factor of 10 [18,23,30]. They synthesize and export high molecular mass polysaccharide mucilage to form a water-soluble capsule surrounding the cap periphery [31].
During the renewal of the root cap, exfoliating border cells (BC) are released daily into the rhizosphere [31,32]. BCs are specific secretory cells, with a central vacuole and a large Golgi apparatus [18]. These cells present an intense metabolic activity, and culture experiments show that they are fully viable after their separation from the root cap [33–35]. Furthermore, their protein content and genomic pattern indicate that they are completely distinct from the columella and the lateral root cap [36,37]. Consequently, BC development could correspond to the third root cap differentiation step (for reviews, see [38–40]).
BC separation from the rest of the root tip requires the action of several cell wall degrading enzymes such as pectin methylesterases [32,41–43]. Their secretion into the mucilage synthesized by root cap secretory cells has been suggested with the ageotropic mutant, which does not produce mucilage and does not release BC [44]. Nevertheless, BC remain attached to the root cap until water solubilises the mucilage structure [32]. After their release, these cells can be found isolated or grouped in small clusters along the root [32,37, 45]. Roots from the Brassicaceae family release very peculiar BC named BC-like (BCL) under the form of 3–4 cells organized into a single layer [45]. In Arabidopsis, three mutants involved in BCL formation have been genetically identified. They display alterations in cell wall components or defects in the secretion of cell wall degradating enzymes, which impacts BC formation in the root tip. The first two are the homogalacturonans deficient quasimodo1 and 2 mutants forming only isolated BC. Homogalacturonans belong to the pectin family and are components of the cell wall [46]. It is the presence of the homogalacturonan in Arabidopsis root tip cell wall that prevents BC cells from separating from each others, and that induces BCL formation [46]. The third mutant is deficient for the CEL5, a 1,4-b-glucanases, and it is altered in BCL release [43]. 1,4-b-glucanase are known to hydrolyse internal linkage of 1,4-b-glucan such as cellulose (the most important structural component of cell walls). In addition, other more complex glucan polymers bearing b-1,4 linkages are also found in the cell wall [43].
The number and type of BC released daily depend on the plant family. It is demonstrated that open meristem species produce dramatically more BC than closed meristem: 10,000 cells daily for pine (open meristem species) compared to 10 for tobacco (closed meristem) [20,45,47]. It must be noted that BC control their own production by secreting a root cap initial division inhibitor into their mucilage. Interestingly, when BC are not released from the root tip, root cap initials show in turn a reduction in root cap cells production [48].
2.5 Genetic and hormonal factors controlling root cap generation
Several cell ablation experiments performed in Arabidopsis thaliana [8] and maize [49,50] have shown that the root cell identity is defined by their position. This positional signal is also at the origin of root cap identity. In support of this, any excision of a root tip will be followed by a regeneration of the root cap by de-differentiation/re-differentiation of the cells located at the root extremity [51,52].
The plant hormone auxin has been shown to play a major role in root patterning (for a recent review, see [53]). Auxin distribution in the root is controlled by tissue specific localization and activity of auxin efflux (PIN) carriers [54–56]. This hormone forms an apico-basal gradient with its maximum centred on the initial columella layer. A combination of genetic and pharmacological experiments affecting auxin transport or its perception have shown the importance of this hormone gradient for the acquisition of the different cellular identities along the apico-basal root axis, including for the root cap [54,55,58,59]. The loss of auxin accumulation in root cap initials induces the apparition of additional columella layers [54].
The root cap formation appears modulated by auxin via the action of auxin responsive factors (ARF). These factors are acting on two types of transcription factors (AP2/ERF and NAC) that control, respectively, the root cap differentiation, and the root cap cell division.
The root cap differentiation is indeed controlled by the four paralogous AP2/ERF-type transcription factors, PLETHORA1 to 3 (PLT1, PLT2, PLT3, [60]) and BABY BOUM (BBM, [61]). This is well illustrated by the fact that the ectopic expression of PLT genes in the embryo results in the ectopic RAM and root cap differentiation [60,62]. All the transcripts for these genes are induced by auxin (through ARF 5 and 7), and their strong expression is tightly correlated with the auxin maximum observed at the root tip [60]. Furthermore, the strong decrease of PIN genes expression in plt mutants suggests a self-reinforcing loop between the auxin flow and PLT expression [55,62]. Wang et al. revealed that, in Arabidopsis, the root cap identity is also under a microRNA regulation by demonstrating that ARFs 10 and 16 are repressed by miR160 ectopic expression. In accordance with their specific expression pattern (in particular ARF 16 expression in the embryo is associated with the hypophyse at early heart stage and the first root cap cells), ARF10 and 16 were found essential for root cap identity acquisition and division regulation [63].
The root cap cell division is under the control of the second major type of transcription factors controlled by ARFs, i.e. the NAC domain transcription factors FEZ and SOMBRERO (SMB). FEZ appears required for periclinal division in root cap initials. Such activity is restricted to root cap initials because FEZ activates its own negative regulator (SMB) in daughter cells [64]. Also, in the fez mutant the ectopic activity of SMB in columella promotes division and a second columella initial layer is generated. Nevertheless, none of these genes play a role in root cap identity, which remained unchanged.
Recent studies revealed the involvement of additional phytohormones (among which Brassinosteroids, Auxin and Ethylene) in root cap formation. Many of them simply act on root cap size through their effects on RAM mitotic activity via interlaced pathways (for a review, see [65]). For example, the brassinosteroids stimulate meristematic cell (QC and columella initials) division and specification by maintaining auxin sensitivity and distribution [66]. Recently, two molecular genetic studies shown that gibberellins promote cell proliferation in the root meristem through the repression of cell cycle inhibitors [67]. In another work, genetic and pharmacological experiments in maize and Arabidopsis revealed that ethylene regulates statocytes differentiation [68]. However, unlike auxin, ethylene represses also BC production by decreasing the RAM mitotic activity [45,68,69]. Furthermore, it was demonstrated that the crosstalk between the auxin and ethylene signaling contributes to root cap formation [68,70].
3 The functions of the root cap
3.1 Root growth
It was shown in maize that the root cap is important for the penetration of the root in soil: it reduces friction, thanks to the secretion of mucilage by secretory cells and the release of BC. In fact, the absence of secretion increases by 30–40% the soil resistance to root growth with a contribution at 58% for BC and 42% for mucilage [19,22]. In compacted soils, secretion of vesicles from secretory cells and mucilage production are increased [71].
Recent studies about the impact of auxin carriers on the auxin polar distribution pattern in roots further demonstrated the importance of the root cap regarding the direction of the auxin fluxes [54,55]. It was shown earlier that the genetic or surgical primary root cap ablation inhibits primary root growth, stimulates secondary root initiation, and modifies the root tissue patterning (by altering the auxin distribution in the root tip) [72,73].
3.2 Interaction with the rhizosphere
The root cap, due to its location at the tip of the growing root, is a major site of interaction with the rhizosphere. During the renewal of the root cap, exfoliating BC are released into the rhizosphere. These cells remain viable for several days and have the capacity to produce various chemicals altering drastically the relationship between the plant and its surrounding microflora such as, for example, the attraction and immobilization of nematodes, the repealing of bacteria or the decoy of fungi (for reviews, see [74,75]).
In Arabidopsis seedlings, BCL produce, along with the root cap, specific cell wall polysaccharide molecules involved in the interaction with rhizobacteria [45]. Finally, the proteolycally degradation of the root cap secretome abolishes the root tip protection from infection, showing the importance of these secretions [76].
3.3 Anchorage and soil exploration
The root growth response to gravity has been studied since the 19th century (for a review, see [77]). It is now well established that the main site of gravity perception corresponds to the root cap (Fig. 3). However, alternative sites contribute also (although to a lesser extend) to the gravity response, as demonstrated by physiological experiments performed on maize showing that gravistimulated roots produced curvature while the root cap was oriented vertically [78]. In the specific case of the root cap, laser ablation experiments demonstrated that gravity perception occurs in the statocytes (described in Section 2) [79]. The active role of starch in this phenomenon has been demonstrated by the use of mutants altered in its synthesis or accumulation. For example, the starch deficient mutant pgm1 and the starch overaccumulating mutant sex1 demonstrated a lack of normal response and an increased sensibility to gravistimulation, respectively [80,81]. The mechanism converting the gravity force into a biochemical signal remains largely unknown. However, according to Leitz et al. [82], gravity sensing results from the conversion of the gravitational potential energy into a biochemical signal, through the sedimentation of statoliths that displaces the cytosol and deforms the ER membranes, and the rapid release of kinetic energy of the sedimenting statoliths from the ER to activate mechanosensitives sites within the columella cells.
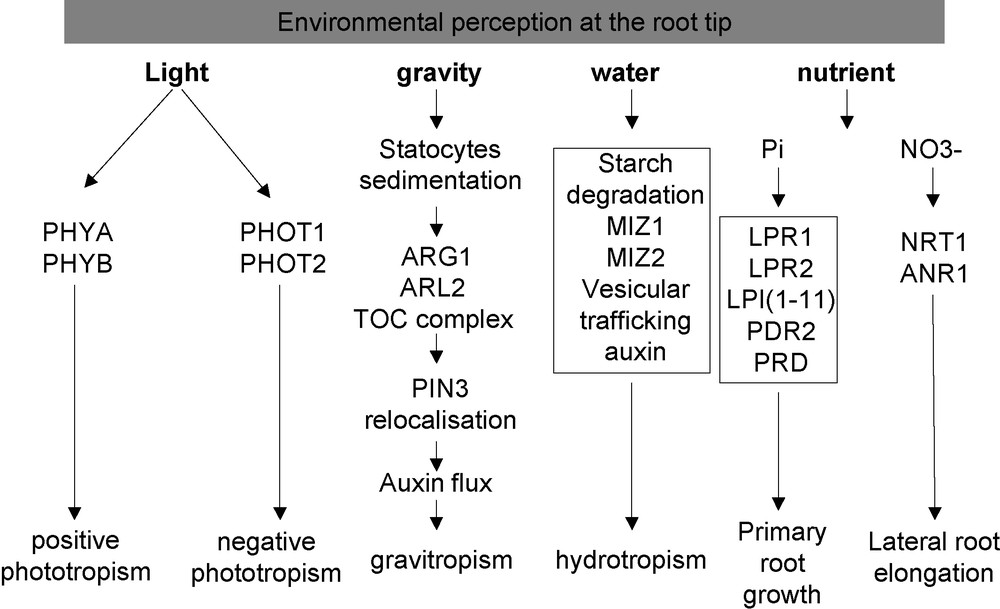
Overview of the environmental perception by the root tip.
The auxin hormone plays an important role for the signal transmission and the growth modification induced by gravitropism changes. Upon gravity stimulation, the auxin efflux facilitator PIN3, localized in columella cells, is rapidly relocalized laterally to the lower side of statocytes [54]. This relocalization requires two proteins specifically expressed in root statocytes: ARG1, a peripheral membrane protein, and its paralog ARL2 [83]. Arabidopsis arg1 and arl2 mutants were the first identified lines affecting gravitropism without altering starch accumulation. Similarly, an enhancer screen identified the mar1 and mar2 mutations that exacerbate the root gravitropic defect of the arg1-2 parental line without affecting amyloplasts morphology [84]. The mar1 and mar2 mutations affect, respectively, the TOC75-III and the TOC132 genes, which are encoding components of the Translocation of Outer Membrane of Chloroplasts (TOC) complex. The known role of the TOC complex is to transport nuclear encoded proteins inside the plastids [85]. It must be noted that in the mar2, arg1 double mutant, the amyloplast morphology and the starch sedimentation keep wild type characteristics. This observation suggests that the TOC complex participates in later steps of the gravity response.
Phototropism of the root tip is also one of the oldest known plant growth responses, and has been studied since the 19th century (for review, see [86]). The ecological and physiological importance of root phototropism is not well understood (Fig. 3). It might reduce exposure to desiccation stress by increasing root efficiency to grow deep into the soil right after germination [87].
Due to the similarity between their associated effects, the specific roles of gravitropism and phototropism were difficult to analyze separately. However, the recent use of gravitropism-defective mutants allowed the identification of specific elements of the phototropism (for a review, see [88]). In Arabidopsis roots, blue light is perceived by the PHOT1 and PHOT2 phototropins that mediate negative phototropism [89,90]. According to genetic data, PHOT1 appears as the main receptor for blue light in roots. It is expressed in the cortical cells of the root elongation zone [91] and its expression is higher at a shallow soil depth, where blue light penetration is the greatest [87].
In maize, the decapped roots failed to develop phototropic curvature. In addition, the use of an optical fibre to illuminate specific regions of the root system revealed that the maize root cap is the site of photoperception for blue light, and that its stimulation induces negative phototropism of the root [92]. The mechanisms of blue light perception in maize root caps remain currently mysterious: do they involve PHOT analogues located in maize root caps, or do they involve other photoreceptors? Furthermore, the phytochrome A or B (PHY A, PHY B) and the cryptochromes CRY1 and CRY2 are also known to be able to perceive blue light and could act in this signalling pathway [93] (Fig. 3).
Recently, a positive red light mediated root phototropism has been reported in Arabidopsis [94], but its role remains elusive. It is mediated by PHY A, expressed in the root cap [95], and PHY B, but its effect is barely detectable and appears weaker than the blue light tropism [96]. Interestingly, the PHYA and PHYB role in positive phototropism strongly interferes with the gravistimulation response [97]. The double mutant phyA, phyB shows indeed reduced rates of gravitropic curvature after gravistimulation. It seems that PKS1, a phytochrome kinase [98,99], provides a complex link between phototropism and gravitropism [100]. In support of this, it was shown that the pks1 mutant is impaired in the negative phototropic response and develops an enhanced gravitropic response.
3.4 Water uptake
The plant root ability to sense and grow towards zones of better water availability (positive hydrotropism) has been demonstrated at the beginning of the 19th century (for a review, see [101]). As in the case of phototropism, the gravitropic response strongly interferes with hydrotropism (Fig. 3). The moisture gradient and the gravity force are both detected in the root tip, and they both trigger a growth response in the elongation zone. Interestingly, the starch content of statocytes decreases during the hydrotropic response, which in turn reduces the amplitude of the root gravitropism [102]. Accordingly, the Arabidopsis nhr1 mutant (not cloned yet), which contains bigger amyloplasts in the root cap, displays a negative hydrotropism and shows a faster positive gravitropic response [103]. One proposed hypothesis is that the starch degradation could be targeted by the osmotic stress, and serve as a metabolisable and osmotically active sugar reserve released during water stress [104].
Two actors for the moisture gradient response have been recently identified by mean of mutant screening: MIZ1 and MIZ2. The study of the ahydrotropic mutant miz permits one to show that the hydrotropism also interacts with phototropism. miz1 shows a more moderate curvature in response to light stimuli than the wild type, without affecting its response to gravistimulation [105]. This observation is correlated with the fact that the miz1 mutation does not modify starch degradation in response to hydrotropism. The MIZ1 gene has been cloned, and it encodes for an unknown protein. It is expressed in columella cells, and MIZ1 mRNA seems to be up-regulated immediately after exposure to osmotic or salt stress [105]. Interestingly, MIZ2 is in fact GNOM, a protein encoding a guanine-nucleotide exchange factor for ADP-ribosylation factor-type G, and it is required for the polar auxin transport through vesicular trafficking [106]. Unexpectedly, the leaky miz2 mutation is defective only in hydrotropism (i.e. not in gravitropism). It also does not present developmental defects like other previously identified gnom mutants. This suggests that vesicular trafficking is crucial for root hydrotropism [107].
The involvement of hormones in hydrotropism is more complex. ABA acts as a critical regulator of tropic responses and plays a role in starch accumulation [104]. The use of drugs revealed that influx or efflux of auxin had no effects on a moisture gradient response. However, the application of p-chlorophenoxyisobutylacetic acid, an inhibitor of auxin response, reduced the root curvature by half in response to a humidity gradient [108]. These observations suggest a novel role of auxin response in hydrotropism through is own response and not through its polar transport.
3.5 Nutrient uptake
Plants have the ability to sense and adapt their root architecture to nutrient availability (Fig. 3; for a review, see [109]). During phosphate starvation, the primary root stops growing and numerous lateral roots are induced [110,111]. The contact of the root tip with the low-Pi media is required for this phenotype, demonstrating two important points: first, that the phosphate starvation status of the whole root system is not the driving mechanism for this response [112], and second, that the root tip plays a specific role in this phenomenon. Several elements of this signalling pathway have been genetically identified by screening for mutants showing a root growth insensitivity, (lpi mutants [113]), or hypersensitivity (pdr2 mutant [114]) to a low phosphate medium. A quantitative genetic analysis, based on the natural variation present between two Arabidopsis ecotypes, identified LPR1 as a major QTL controlling the primary root length in response to Pi starvation [115]. Until now, only LPR1 and PDR2 have been cloned, and they encode a multicopper oxidase [112] and a type 5 P-ATPase [116], respectively. LPR1 belongs to a multigenic family with only one closely related paralog, LPR2. Interestingly, the lpr1, lpr2 double mutation suppresses the hypersensitive phenotype conferred by the pdr2 mutation, suggesting that the LPR1, LPR2 and PDR2 functions are part of the same pathway [116]. Their mode of action remains elusive and complex, since several conditions (such as pH, iron and phosphate contents) can also modulate the root growth response. It remains, nonetheless, clear that the root cap (with the involvement of LPR1 in particular) represents the main location required for triggering the growth response to low phosphate [112]. More recently, it was shown that mutants in the AINTEGUMENTAL-like gene PRD, which is expressed in the root tip, exhibit a mild reduction in the primary and lateral root development under Pi starvation [117]. This provides another line of evidence for the importance of the root apex regarding phosphate perception.
The root tip appears also involved in the root architecture response to nitrate [118], another major macronutriment (Fig. 3; for review see [119]). The main effect of this ion is to locally stimulate lateral root growth. In parallel, it counter-acts the repressive activity of glutamate on the root apical meristem activity. This pathway involves the ANR1 MADS box transcription factor [120], as revealed by the study of ANR1 underexpressing lines, which do not develop any lateral root elongation response in presence of high NO3−.
The dual affinity NO3− transporter NRT1.1 (CHL1) is another major regulator of this pathway. In the chl1 mutant (loss of function), the lateral root elongation in response to local NO3- is reduced and the ANR1 expression is strongly reduced [121]. The phosphorylation and the dephosphorylation of the Tyrosine 101 from the CHL1 protein lead respectively to a high and low affinity transporter [122]. Interestingly, the phosphorylation state of this tyrosine also modulates the NRT1.1 sensitivity towards the nitrate signal [123]. The identification of mutants decoupling the activity of nitrate transport and nitrate sensing confirmed the view that NRT1.1 has a dual function, and behaves as a transceptor [123]. In addition, the strong expression of NRT1.1 in the root tip [121,124] reinforces the view that the root tip represents a major zone for sensing the root external environment.
4 Conclusion
A striking point illustrated by this article is the identification of many environmental sensing pathways in the root tip (Fig. 3). We have underlined the role of the root cap as an important sensing site for external signals. It has also highlighted the fact that plant tropisms interact strongly to optimize the root system architecture according to the soil environment, modulating water and nutrient uptake [125]. Most of the external signals affect the root growth and development. Nevertheless, for each of these signalling pathways, an important gap remains between the sensing mechanisms (for which some actors have been unravelled) and the final modifications affecting the root architecture.
Acknowledgments
We want to thank Dr Hélène Javot for the corrections and comments provided. We are grateful to Géremy Clair for his help in the realisation of the Fig. 2.


