1 Introduction
Plant growth and development is greatly affected by the regulation of water uptake and transport across cellular membranes and tissues. However, under natural conditions, plants are exposed to various soil stress conditions including salinity, drought, temperature extremes, nutrient deficiency, heavy metal toxicity and so on, most of which directly influence hydraulic conductivity (Lpr) and induce an accumulation of reactive oxygen species in root tissues [1]. Under such conditions, plants maintain their osmotic and ion homeostasis by absorption, transportation and compartmentation of water and solutes. Osmotic water transport across membranes is mediated by water channel proteins known as major intrinsic proteins (MIPs), precisely aquaporins (AQPs). The permeability of membranes is actively controlled not only by the regulation of the amount of different MIPs present but also in some cases by phosphorylation/dephosphorylation of the channels, cytosolic calcium, and cytosolic proton, [2,3]. AQPs are characterized by six membrane-spanning α-helices and both C- and N-termini facing the cytosol [4], and two highly conserved loops with the amino acid motifs of asparagine-proline-alanine (NPA).
In recent years, quite a large number of plant MIP genes have been identified. For instance, in Arabidopsis (Arabidopsis thaliana) 35 MIP genes have been identified [2]. Similarly, in maize (Zea mays) and rice (Oryza sativa) 33 genes encoding the MIPs have been reported [5,6]. Plant MIPs are classified into at least four subfamilies based on their sequence similarity: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs) and small basic intrinsic proteins (SIPs) [2]. Recently, a fifth uncharacterized group of MIPs (XIPs) has been identified in Physcomitrella patens [7] and Gossypium hirsutum [8]. Members of the PIP and TIP subfamilies are abundant in the plasma membrane and in the vacuolar membrane (tonoplast), respectively. The third subfamily has been named NIPs [9]. NOD26, the first identified member of this subfamily, is located in the peribacteroid membrane of nitrogen-fixating symbiosomes in root nodules of soybean (Glycine max) [10]. Three genes belonging to the NIPs subfamily previously reported in Arabidopsis (AtNIP5;1, AtNIP2;1) and rice (OsLsi1) are localized in the plasma membrane [11–13]. The fourth subfamily was named small basic intrinsic proteins (SIPs). Neither the substrate specificity nor the intracellular localization of SIPs is well understood [2,14]. However, it has been reported that proteins of three Arabidopsis SIPs (AtSIP1;1, AtSIP1;2 and AtSIP2;1) are localized in the ER, but were not detected in the plasma membrane or the vacuolar membrane [15].
The functions of plant MIPs have been suggested from purified plant membranes by using inhibitors, heterologous expression in oocytes, complementation studies in yeast (Saccharomyces cerevisiae) mutants [16–18], or overexpression in plants. Accordingly, several MIP isoforms are known to function as water channels [16,19,20]. Besides water MIPs are known to facilitate the transport of glycerol, CO2, NH4+/NH3, urea, B, Si and H2O2, arsenic, antimony and lactic acid [11–13,16,18,21–29].
Several studies have shown that abiotic stresses such as nutrient deprivation, water deficit, salinity and waterlogging reduce root hydraulic conductivity [30], and this response in most of the cases is related to a decrease in MIPs activity. Therefore, we were interested in studying the effect of abiotic stresses on the expression of MIPs in barley. It has been indicated that expression of MIPs genes is influenced by drought, salt, metals and hormone treatment, and expression is differentially regulated in various plant organs [6,30–33]. In Arabidopsis, the expression of MIP genes in different organs in response to cold, salt, drought and ABA treatment has been determined by using a semi-quantitative slot blot analysis and quantitative real-time RT-PCR [32–34]. Most of the studies showed that several AQPs were predominantly expressed in one organ and that many were markedly up or downregulated under the different stress conditions. Analysis of 33 rice MIP gene expression at different growth stages and in different plant organs also showed that gene expression varied with plant organ and growth stage [6].
Heavy metals are toxic to higher plants by causing oxidative stress, displacing other essential metals in plant pigments or enzymes, disrupting the function of these molecules and of many metabolic processes such as electron transport chain, and finally reducing growth and yield [35–40]. Although heavy metals are known to inhibit most MIPs, it remained to be elucidated whether the effect of such metals involves transcriptional or post-translational modification of the proteins. Heavy metals such as Cd, Cu and Hg are known to modulate gene expression and function of MIPs in various plant species. For example, in Pisum sativum reduction of Lpr by HgCl2 treatment was accompanied by an increase in the expression of PsPIP2;1, suggesting that the increase in PsPIP2;1 might compensate for the AQPs blocked by Hg [41], whereas in Populus deltoides roots subjected to copper stress genes encoding plasmalema (PIP) and tonoplast (TIP) AQPs were downregulated under Cu application [42]. Similarly, in Solanum torvum 15 tags annotated to Arabidopsis TIPs, PIPs, SIPs, and a NIP, were significantly downregulated, and one and two tags annotated to a TIP and PIPs, respectively, were upregulated in response to Cd stress. In most cases, these transcriptional changes were rapid and occurred within 3 h [43]. Inhibition of aquaporin function due to direct interaction of heavy metal with AQPs has also been suggested. Previous studies in wheat (Triticum aestivum) and tomato (Solanum lycopersicum) showed that a 10 minutes exposure to HgCl2 significantly reduced Lp indicating the interaction of Hg with the existing water channel proteins [44–45]. Similarly, it has been shown that Cd directly binds with AtPIP2;1 and inhibits its water transport activity in reconstituted liposomes [3].
Mineral nutrient deficiency is another important constraint dictating plant growth and development. The availability of major nutrients such as nitrate or phosphate significantly influences root hydraulics [46–48]. The deficiency of N, P and S is reported to induce a progressive drop in Lpr and influence plant water status, whereas a supply of nutrient results in its rapid upregulation [1,17,49–50]. However, the effect of nutrient limitation on MIPs gene expression has not been well studied.
In one of the most important crops barley, a number of putative MIP isoforms (10 PIP, 8 TIP, 3 NIP, and a SIP gene) have been identified from the high homology contiguous sequences predicted from EST database (Fig. 1). Only a few of the MIPs have been isolated, while most MIPs have not been identified and their physiological roles under various abiotic stress conditions are not known. Hence, the purpose of the current study was to clone, characterize gene expression under various abiotic stresses (salt, heavy metals and nutrient deficiency) and phytohormones (ABA and GA), and analyze the physiological function of the MIPs gene products by heterologous expression in oocytes.
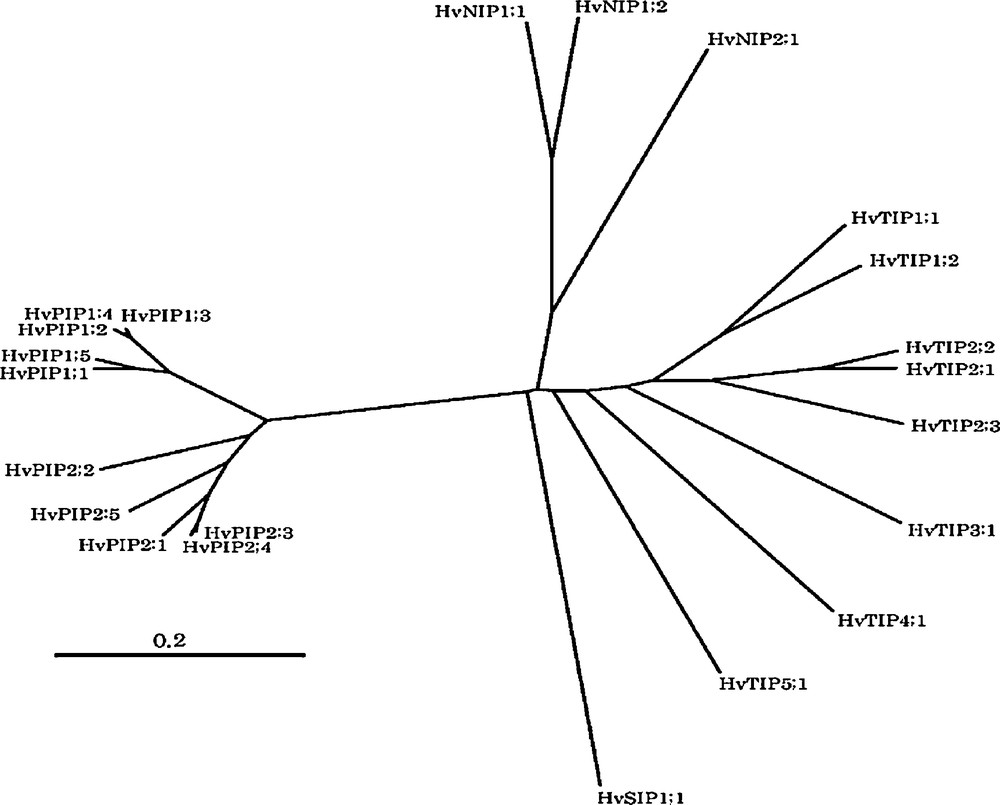
Phylogenetic tree of barley MIPs based on contiguous sequences predicted from EST database by HarvEST software (http://harvest.ucr.edu). Registered genes and mostly full-length contigues were used to generate the phylogenetic tree. Amino acid alignment and the phylogenetic tree were generated by CLUSTALW (1.81) at http://align.genome.jp with unrooted N-J tree method. The scale bar represents the evolutionary distance, expressed as the number of substitutions per amino acid. Following is accession no. of registered genes: HvPIP1;1, AB286964; HvPIP1;2, AB275278; HvPIP1;3, AB009308; HvPIP1;4, AB275279; HvPIP1;5, AB009309; HvPIP2;1, AB009307; HvPIP2;3, AB275280; HvPIP2;4, AB219525; HvTIP1;1, AB540221; HvTIP1;2, AB540226:, HvTIP2;1, AB292847; HvTIP2;2, AB540223; HvTIP2;3, AB261102; HvTIP3;1, AB540228; HvTIP4;1, AB540225; HvTIP5;1, AB540227; HvNIP1;1, AB540230; HvNIP1;2, AB540231; HvNIP2;1, AB292848).
2 Materials and methods
2.1 Plant growth and treatments
Seeds of barley (Hordeum vulgare L. cv. haruna-nijo), a moderately salt tolerant cultivar, were obtained from the Barley Germplasm Center of the Institute for Plant Science and Resources. The seeds were surface sterilized with 10% H2O2 for 10 min. After rinsing five times with deionized water, the seeds were soaked overnight with continuous aeration. The germinating seeds were further rinsed and spread on a mesh which was placed on top of a 3.5 L pot filled with 0.25 mM CaSO4.2H2O solution. The seeds were covered with moist paper towel, which was immersed in the Ca-solution to protect the germinating seeds from desiccation. The pots were then covered with aluminium foil and the seedlings were grown in a dark and ventilated room at 25 °C. After two days, the seedlings were transferred to an aerated nutrient solution containing 4 mM KNO3, 1 mM NaNO3, 1 mM NaH2PO4.2H2O, 1 mM CaCl2.2H2O, 1 mM MgSO4.7H2O, and micronutrients (1 ppm Fe, 0.5 ppm B, 0.5 ppm Mn, 0.05 ppm Zn, 0.02 ppm Cu, and 0.01 ppm Mo). The pH of the nutrient solution was adjusted to 5.5 with 0.2 N NaOH.
To study the effect of salt, ABA, GA and heavy metals (Cd, Cu, Hg, Zn and Cr) on gene expression, five-day-old seedlings were transferred to the nutrient solution containing 100 mM NaCl, 0.1 mM ABA, 1 mM GA or 200 μM of metals (CdCl2, CuCl2, HgCl2, ZnCl2 or CrCl2). After 24 h, two representative roots and shoots samples were taken from each treatment for RNA extraction.
It has been reported that the deficiency of major nutrients such as N, P and S decreases Lpr [17,50]. However, it remained unclear if such stimuli modulate the expression of MIPs. Hence, we investigated if these nutrients affect MIPs gene expression. For N-deficiency treatment, seedlings cultured in 0.25 mM CaSO4.2H2O solution at dark were transferred to a nutrient solution in which KNO3 and NaNO3 were replaced by KCl and KH2PO4, respectively. For P-deficiency treatment, NaH2PO4.2H2O was omitted from the nutrient solution. The plants were then transferred to a growth chamber (MLR-351H, Sanyo, Osaka, Japan) at a cycle of 14 h day and 10 h night continuously at 25 °C, and light intensity of 150 μmol m−2 s−1 using Sanyo FL40SS.W/37 fluorescent lamps. Roots and shoots samples were taken at the same time of the day for RNA isolation every 3 days. After six days of either N or P-deficiency, the plants were transferred to a complete nutrient solution and samples were taken three days after recovery.
2.2 Determination of rate of transpiration
Since several fundamental plant processes such are transpiration are greatly affected by water absorption, we studied the effect of heavy metals on water uptake by measuring the rate of transpiration from 6-day-old seedlings that were previously exposed to 200 μM each of CdCl2, CuCl2, HgCl2, ZnCl2 or CrCl2 for 24 h. For this purpose, seedlings were carefully placed between two halves of silicon rubber on which Vaseline was smeared. The seedlings were then transferred to glass vials containing 25 mL of Artificial Pond Water (APW, 0.1 mM of CaCl2, NaCl and KCl). The top of the vials was tightly sealed with Vaseline to avoid water loss. The initial weight of the vials containing the seedlings was recorded and the seedlings were incubated at 25 °C for 1 h in the dark. The seedlings were kept under a glass dome in which silica gel was placed to absorb water lost from the seedlings through transpiration. The final weight of the vials was recorded and the decrease from the initial weight was considered as the amount of water loss from the seedlings through transpiration.
2.3 RNA extraction
For gene expression analysis, 0.1 g apical root (0–3 cm) and shoot (about 2–3 cm below the tip of fully expanded leaves) tissues were sampled after the desired treatment and immediately frozen in liquid nitrogen. The frozen plant material was disrupted by mortar and pestle. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Toko Japan) following the manufacturer recommended procedure. For cloning the MIP genes, 1 g apical root tissue was excised and frozen in liquid nitrogen. The plant material was then lysed with Trizol reagent (Invitrogen/Life Technologies, Carlsbad, California, USA). RNA was isolated by a modified acid guanidinium thiocyanate-phenol-chloroform method [51]. The RNA samples were stored at −80 °C for subsequent use.
2.4 Gene cloning
Several genes belonging to the MIPs have been identified based on the high homology contiguous sequences predicted from barley EST database. The current study was planned to clone and characterize the MIPs. For cloning, mRNA was prepared from total RNA using the QuickPrep micro mRNA purification kit (Amersham Bioscience, England, UK). One microgram mRNA was reverse transcribed by the Superscript™ first strand synthesis system for RT-PCR (Invitrogen/Life Technologies). Primers specific to each contiguous sequence (Table 1) were designed to amplify the MIP genes by TaKaRa Ex Taq Kit (TAKARA BIO INC, Shiga, Japan). The PCR products with expected size were purified from gel using Qiaquick Gel Extraction Kit (Qiagen, Tokyo, Japan) and cloned into TOPO TA cloning vector (Invitrogen/Life Technologies). The products were sequenced according to the enzymatic method [52] using an ABI PRISM BigDye terminator cycle sequencing reaction kit, ver. 1.1 (Applied Biosystems, Foster city, CA, USA) with a DNA sequencer 3100-Avant genetic analyzers (Hitachi Ltd., Tokyo, Japan). The nucleotide and deduced amino acid sequence were analyzed by GENETYX software (GENETYX CORPORATION, Tokyo, Japan).
Primers used for cloning and gene expression analysis by real-time RT-PCR.
| Name | GenBank accession number | Primers | Partial | Full length | bp | Real-time PCR | bp |
| HvTIP1;2 | Sense Antisense |
5’-TTCGTGGGCGGCAACATCAG-3’ 5’-CAGGATGTTGGCGCCGAC-3’ |
5’-ACATCAGCCTCCTCAAGGC-3’ 5’-CGATGAAGCCGATGGCGG-3’ |
288 | |||
| HvTIP2;1 | AB292847 | Sense Antisense |
5’-CACATCACCATCCTCACCGG-3’ 5’-GCCGGCGGCGAGGATGTT-3’ |
5’-GCATAAGCCCTCTTTGAGTGT-3’ 5’-CGGCCGAACATTCTGAAAAAG-3’ |
744 | 5’-GCGATGGTGCCGAGGGAT-3’ 5’-CACCGGGATCTTCTACTGA-3’ |
286 |
| HvTIP2;2 | Sense Antisense |
5’-ACCATCCTCACCGGGCTCT-3’ 5’-ACGATGAAGCCGATCGCCAT-3’ |
5’-GCGATGCCGAGGGAC-3’ 5’-CACCGGGCTATTCTACTGG-3’ |
254 | |||
| HvTIP2;3 | AB261102 | Sense Antisense |
5’-CTCGGCGGTCAGATCACCA-3’ 5’-GCAACGAGGATGTTGGCC-3’ |
5’-GACGAAGATTTCATCTCTCATCC-3’ 5’-GAATCGCATAAGGAAACTCAAC-3’ |
744 | 5’-CTACTGGGTTGCGCAGCTC-3’ 5’-GTGCCGAGGGATCCCTTC-3’ |
284 |
| HvTIP4;1 | Sense Antisense | 5’-TCCGGGCGGTGCTGTACG-3’ 5’-TGGTGTTGGCACCGACAATA-3’ | 5’-CACCGACAATAAGGCCGGT-3’ 5’-CGGTGCTGTACGTGGTGG-3’ |
265 | |||
| HvNIP2;1 | AB292848 | Sense Antisense |
5’-TACGGCGCCGACGTGA-3’ 5’-GTCGTCCCCAGCACCGTGA-3’ | 5’-TCACACGCCTCATCTTCTCTT-3’ 5’-TTGCAGGAAAAGCAAAGCTC-3’ |
831 | 5’-TGACGCGCGTCTCGCAGC-3’ 5’-CCAGCACCGTGATCGGGT-3’ |
258 |
| HvPIP2;1 | AB219366 | Sense Antisense |
5’-GCTAGCTTAGCAATGGCCAAGGAC-3’ 5’-AGAGCGAGCAAAAGAAAAGCACAC-3’ |
858 | 5’-GCTAGCTTAGCAATGGCCAAGGAC-3’ 5’-GTCGGACTGGTGCTTGTACC-3’ |
216 | |
| Name | Primer | Primers for oocyte expression | |||||
| HvTIP2;1 | Sense Antisense |
5’-CCGAGATCTATGGTGAAGCTCGCATTC-3’ 5’-CCGGAGATCTCGCGTAGTCCTGGTCCGC-3’ |
|||||
| HvTIP2;3 | Sense Antisense |
5’-TCCAGATCTATGCCAGGCTCCATCGCC-3’ 5’-GCGAGATCTGTAGTCGTTGCTGGCGAC-3’ |
|||||
| HvNIP2;1 | Sense Antisense |
5’-CGGAGATCTATGTCGGTGACTTCCAAC-’3’ 5’-GGCAGATCTGACATGGTCGAACTCGGTC -3’ |
2.5 Analysis of gene expression
Gene expression was studied by the quantitative real-time RT-PCR techniques. First strand cDNA synthesis was performed in a 20 μL reaction that contained 2 μg of total RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). The PCR reactions were initiated by adding 1 μL of the first-strand cDNA to 50 μL mixture containing 1 × Power SYBG PCR master mixture (Applied Biosystems), 200 nM sense, and antisense primers (Table 1). Quantitative PCR was performed on the 7300 Real Time PCR system (Applied Biosystems) with PCR conditions of 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 56 °C for 30 s, and 60 °C for 1 min. For absolute quantification of transcript copy numbers, the partial sequence of the genes was cloned into the TOPO TA cloning vector (Invitrogen/Life Technologies). The vector containing the genes was linearized by SacI that has a unique restriction site. Complementary RNA was generated using the MEGA Script® T7 Kit (Ambion, Austin, TX, USA). A serial dilution of the cRNA of known copy number (105 to 1010, calculated from the size and molecular weight of cRNA) was reverse transcribed along with the samples. The first strand cDNA from the cRNA was used as reference to determine the absolute transcript copy number of the MIP genes in a given microgram of total RNA of the samples.
2.6 Test of water channel activity in Xenopus oocytes
For functional analysis, the coding regions of the HvTIP2;1, HvTIP2;3 and HvNIP2;1 were amplified with primers including BGlII restriction sites (Table 1). After digesting with the BGlII, the fragments were inserted into the BglII site of the pXBG-ev1 vector, that is a pSP64T-derived Bluescript type vector into which oocytes β-globin 5′ and 3′ untranslated regions have been inserted [53]. The construct was linearized with NotI and cRNA was synthesized using the mMESSEGE mMACHINE T3 in vitro transcription kit (Ambion), the synthesized cRNA was suspended in RNase free water. Uniformly sized oocytes were obtained from ovarian lobes isolated from anesthetized adult female frogs as described by earlier [54]. The oocytes were incubated overnight at 20 °C in modified Barth's saline [MBS; 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 15 mM Tris-HCl (pH 7.6), 0.3 mM Ca (NO3)2.4H2O, 0.41 mM CaCl2.4H2O, 0.82 mM MgSO4.7H2O, 10 mg mL−1 sodium penicillin, and 10 mg mL−1 streptomycin sulfate]. The oocytes were then injected with 50 nL (∼50 μg) of cRNA solution of the MIPs, cRNA of HvPIP2;1 (positive control, [54] or with 50 nL MilliQ water (negative control). A day after the injection of cRNA, oocytes were transferred from control MBS of 200 mOsm (osmin) to 5-fold diluted MBS of 40 mOsm (osmout). Osmotic water permeability in the oocytes was determined by a swelling assay. The time course changes in oocyte volume after hypotonic treatment were exported to a computer and analyzed. Osmotic water permeability (Pf) was calculated from osmotic swelling data, as:
Pf = [V0 × d(V/V0)/dt]/[S × Vw × (osmin–osmout)],where initial oocyte volume (V0 = 9 × 10−4 cm3), relative volume (V/V0), surface area of oocyte (S = 0.045 cm2), and the molecular volume of water (Vw = 18 cm mol−1).
2.7 Data analysis
To quantify the transcript copy number, mRNA was isolated from four biological replicates, and at least two independent experiments were conducted. Statistical tests of significance were performed using the PROC GLM procedure in SAS. Following significant F-tests, the Tukey multiple comparison procedure for all treatment pairs was used to separate the means [55]. This was done separately for each gene.
3 Results
3.1 Gene cloning
The cDNAs of six MIP isoforms (HvTIP1;2, HvTIP2;1, HvTIP2;2, HvTIP2;3, HvTIP4;1 and HvNIP2;1) have been cloned from the high homology contiguous sequences predicted from barley EST database using gene-specific primers (Fig. 1). The ORF of three genes (HvTIP2;1, HvTIP2;3 and HvNIP2;1) has been isolated. The HvTIP2;1 and HvTIP2;3 have 70% amino acid homology to each other and 35% homology with HvNIP2;1. Moreover, the HvTIP2;1 and HvTIP2;3 showed about 70 and 95% amino acid similarity with TaTIP2;1, respectively. Similarly, the HvNIP2;1 shares 85 and 75% amino acid similarity with ZmNIP2; 2 and OsLsi1, respectively (Fig. 2). All the three barley MIPs posses six transmembrane spanning domains and the two highly conserved motifs of Asp-Pro-Ala (NPA), which is a typical characteristic of aquaporin gene family.

Multiple sequence alignment of MIPs from barley (HvTIP2;1, AB292847; HvTIP2;3, AB261102; HvNIP2;1, AB292848 and HvPIP2;1, AB009307), wheat (TaTIP2;1, AY525639), maize (ZmNIP2;2, AF326485) and rice (OsLsi1, AB222272). Alignments were carried out using CLUSTALW (http://clustalw.genome.jp). Identical amino acids and similar amino acids were indicated by dark shading and light shading, respectively, using BOXSHADE software (http://www.ch.embnet.org/software/BOX_form.html). The two NPA motifs are indicated by the rectangles.
3.2 Salt, ABA and GA differentially regulate expression of MIP genes
To investigate the effect of salt, ABA and GA on the expression of MIP genes, the partial sequences of seven MIP genes including HvPIP2;1 (a barley aquaporin reported to facilitate water transport) [54], were cloned and gene expression was studied by the quantitative real-time RT-PCR using gene-specific primers. Plants were exposed to 100 mM NaCl, 0.1 mM ABA and 1 mM GA for 24 h and the transcript copy number was quantified from total RNA, and data on transcript abundance are shown in Table 2. In roots, salt stress increased the transcripts of HvTIP1;2 and HvPIP2;1 but transcripts of HvNIP2;1 were significantly decreased. Exogenous application of ABA significantly enhanced expression of HvTIP2;1, HvTIP4;1, HvNIP2;1 and HvPIP2;1. Also GA increased expression of HvTIP1;2, HvTIP2;1 and HvPIP2;1, whereas in shoots, salt stress enhanced expression of HvTIP2;1 and HvNIP2;1 but did not significantly affect expression of other MIPs (Table 2). Abscisic acid enhanced expression of HvNIP2;1 but decreased that of HvTIP1;2 and HvPIP2;1. Gebberellic acid upregulated HvTIP4;1 and HvNIP2;1 but suppressed expression of HvPIP2;1 significantly.
The effect of salt and phytohormones on copy number of MIP transcripts. Plants were treated with 100 mM NaCl, 0.1 mM ABA or 1 mM GA for 24 h. Total RNA was isolated and transcript copy number was analyzed by a quantitative real-time RT-PCR as described in Materials and methods.
| Tissue | Treatment | TIP1;2** | TIP2;1* | TIP2;2** | TIP2;3*** | TIP4;1** | NIP2;1** | PIP2;1** |
| Root | Control | 4.7 ± 0.1c | 2.1 ± 0.1b | 38.3 ± 1.0a | 1.1 ± 0.1a | 4.6 ± 0.0bc | 3.5 ± 0.1b | 3.2 ± 0.1c |
| NaCl | 8.6 ± 0.1a | 2.8 ± 0.1b | 39.4 ± 0.2a | 0.7 ± 0.1a | 4.4 ± 0.1c | 0.5 ± 0.2c | 6.8 ± 0.4b | |
| ABA | 6.1 ± 0.7bc | 4.2 ± 0.3a | 25.3 ± 0.5c | 1.5 ± 0.5a | 28.8 ± 4.2a | 4.6 ± 0.2a | 4.0 ± 0.1b | |
| GA | 7.1 ± 0.6ab | 3.9 ± 0.3a | 32.7 ± 0.8b | 0.7 ± 0.2a | 13.5 ± 0.7b | 3.5 ± 0.4b | 13.5 ± 2.1a | |
| Shoot | Control | 7.3 ± 0.4a | 5.3 ± 0.3b | 24.6 ± 7.0a | 12.1 ± 4.2ab | 25.7 ± 0.7b | 1.6 ± 0.26c | 12.0 ± 0.7a |
| NaCl | 7.1 ± 0.8a | 7.0 ± 0.3a | 16.0 ± 4.7a | 13.9 ± 0.9a | 28.9 ± 2.8b | 5.1 ± 0.2a | 10.1 ± 0.6a | |
| ABA | 3.5 ± 0.3b | 4.2 ± 0.4b | 23.3 ± 2.4a | 3.9 ± 0.5b | 23.9 ± 0.3b | 3.0 ± 0.3b | 2.8 ± 0.3c | |
| GA | 6.1 ± 0.2a | 4.6 ± 0.2b | 30.9 ± 0.8a | 8.0 ± 1.3ab | 38.7 ± 1.5a | 5.0 ± 0.3a | 6.4 ± 0.5b |
3.3 Heavy metals modulate expression of MIPs and inhibit water uptake in barley
Since heavy metals such as Hg are known to block water movement by inhibiting the activity of AQPs, we studied the effects of various heavy metals on MIPs gene expression. For this purpose, plants were exposed to 200 mM Cd, Cu, Hg, Zn or Cr for 24 h and absolute transcript copy number was determined. In roots, Cd, Cu, Zn and Cr downregulated expression of HvTIP1;2, HvTIP2;2, HvNIP2;1 and HvPIP2;1 (Table 3). Intriguingly, Hg which is known to inhibit water channels, upregulated expression of HvTIP2;3, HvNIP2;1 and HvPIP2,1 but suppressed transcripts of HvTIP1;2 and HvTIP4;1. Expression of HvTIP4;1 was not affected by Cu, Zn and Cr, while transcripts of HvTIP2;3 were suppressed by Cd, Cu, Zn and Cr. The effect of Cd, Cu and Cr was more pronounced in roots. In shoots, expression of most MIPs was not significantly affected (Table 3) by the metals. Metals (Cd, Hg, Zn and Cr), except Cu, stimulated transcripts of HvTIP2;2 and HvTIP2;3. Moreover, transcripts of HvPIP2;1 were stimulated by Hg by two-folds.
The effect of heavy metals on copy number of MIP transcripts. Plants were treated with 200 μM of the metals for 24 h. Total RNA was isolated and transcript copy number was analyzed by a quantitative real-time RT-PCR as described in the Materials and methods.
| Tissue | Treatment | TIP1;2** | TIP2;1* | TIP2;2** | TIP2;3*** | TIP4;1** | NIP2;1** | PIP2;1** |
| Root | Control | 24.6 ± 1.0a | 6.0 ± 1.9ab | 76.3 ± 2.8a | 31.3 ± 1.9b | 20.7 ± 0.3a | 6.7 ± 0.2b | 10.6 ± 0.6b |
| Cd | 9.04 ± 0.3c | 3.1 ± 0.1b | 15.8 ± 1.0c | 1.0 ± 0.1d | 15.0 ± 1.0bc | 3.5 ± 0.1d | 2.3 ± 0.1d | |
| Cu | 12.6 ± 0.6c | 3.4 ± 0.4b | 16.5 ± 0.4c | 0.6 ± 0.1d | 21.1 ± 0.3a | 2.5 ± 0.1e | 2.9 ± 0.4d | |
| Hg | 20.2 ± 0.5b | 9.6 ± 2.2a | 82.1 ± 14.7a | 48.9 ± 1.3a | 12.8 ± 0.5c | 9.5 ± 0.3a | 12.4 ± 0.6a | |
| Zn | 17.8 ± 0.7b | 4.2 ± 0.6ab | 46.9 ± 1.3b | 10.1 ± 0.4c | 16.5 ± 1.0abc | 4.5 ± 0.1c | 5.1 ± 0.3c | |
| Cr | 9.9 ± 0.7c | 3.8 ± 0.2b | 22.5 ± 1.7bc | 2.7 ± 0.5d | 19.2 ± 1.9ab | 2.0 ± 0.2e | 2.6 ± 0.3c | |
| Shoot | Control | 8.3 ± 0.2a | 3.7 ± 0.6a | 16.7 ± 0.1b | 3.5 ± 0.0d | 19.0 ± 0.5ab | 6.5 ± 0.4ab | 1.4 ± 0.1bc |
| Cd | 6.1 ± 0.1dc | 4.3 ± 0.2a | 33.6 ± 2.3a | 13.2 ± 0.4a | 15.4 ± 0.8bc | 4.5 ± 0.1b | 1.7 ± 0.1bc | |
| Cu | 7.9 ± 0.9abc | 4.0 ± 0.2a | 16.7 ± 1.9b | 2.3 ± 0.6d | 12.2 ± 1.1c | 6.3 ± 1.2ab | 1.2 ± 0.3bc | |
| Hg | 6.4 ± 0.1bdc | 6.9 ± 2.3a | 31.3 ± 0.7a | 11.7 ± 0.8ab | 16.0 ± 0.8abc | 8.6 ± 0.2a | 3.1 ± 0.3a | |
| Zn | 8.0 ± 0.2ab | 4.8 ± 0.7a | 28.4 ± 4.7a | 9.2 ± 1.2bc | 14.5 ± 0.4bc | 4.7 ± 0.5b | 1.9 ± 0.2b | |
| Cr | 5.6 ± 0.2d | 4.6 ± 0.2a | 30.1 ± 0.6a | 7.8 ± 0.5d | 20.5 ± 2.0a | 4.6 ± 0.3b | 0.9 ± 0.1c |
To investigate the effect of heavy metals on water uptake, we measured the rate of transpiration from intact plants that were previously exposed to 200 μM of the metals for 24 h. As presented in Fig. 3, the rate of transpiration was significantly suppressed by metal treatment (p < 0.0001). Mercury inhibited the rate of transpiration by about 45%. Overall, metal treatment decreased the rate of transpiration by 30–45% indicating that the metals may suppress the uptake of water by plant roots.

The effect of 200 μM CdCl2, CuCl2, HgCl2, ZnCl2 or CrCl2 exposure for 24 h on the rate of transpiration. Water loss through transpiration was determined as described in the Materials and methods. Bars represent the average of two independent experiments each with 10 replicates. Error bars indicate SE.
3.4 Nitrogen and phosphorus-deficiency downregulate expression of MIPs in roots
Plants were first exposed to six days of either N or P-deficiency. After six days, the plants were transferred to a complete nutrient solution. RNA was isolated every 3 days for gene expression analysis. Six days of N-deficiency downregulated expression of HvTIP1;2, HvTIP2;1, HvTIP2;2, HvTIP2;3, HvNIP2;1 and HvPIP2;1 in roots (Fig. 4A–G), whereas expression of HvTIP4;1 was upregulated by N-deficiency. Interestingly, expression in the N-deficient plants was restored to control level 3 days after N was supplied. Expression of the MIPs was not markedly affected by N-deficiency in shoots (Fig. 4H–N) but transcripts of HvTIP2;1 and HvNIP2;1 were slightly upregulated. Unlike N-deficiency, three days of P-deficiency increased expression of all MIPs. However, when P-deficiency was prolonged to 6 days, transcript copy number decreased sharply below (for HvTIP1;2 and HvPIP2;1) or to expression level in control plants (for HvTIP2;1, HvTIP2;2 and HvNIP2;1) (Fig. 5A–E). In shoots, 3 days of P-deficiency increased expression of HvTIP2;1, HvTIP2;2 and HvTIP2;3 but decreased expression of HvTIP1;2, HvNIP2;1 and HvPIP2;1 (Fig. 5H–N). In both roots and shoots, gene expression was recovered to control level three days after P was supplied. Overall, the transcript copy number of all the MIPs was more abundant in roots than shoots. Moreover, HvTIP2;3 and HvTIP2;1 have the highest and the lowest copy numbers, respectively.
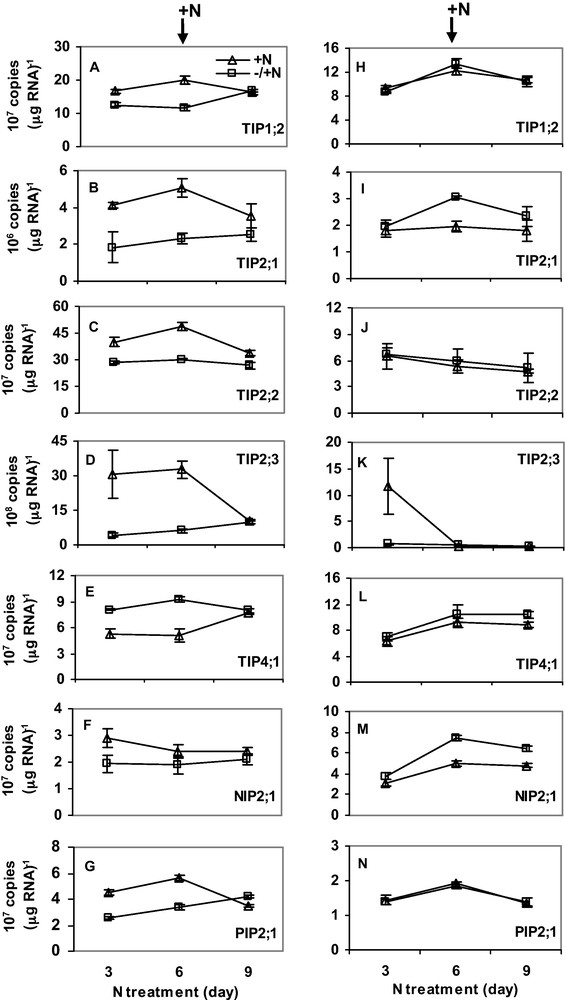
The effect of N-deficiency on the expression of MIPs in roots (A–G) and shoots (H–N). For N-treatment, N supply was resumed six days after deficiency as described in the Materials and methods. RNA was isolated from root tips and the youngest fully expanded leaves at the same time of the day. Gene expression was studied every three days. Curves (–/ + N, open square; + N, open triangle) represent the average of two independent experiments each with four replicates, and error bars indicate SE. Note that the scale of Y-axis in the figures varies depending on the type of gene.
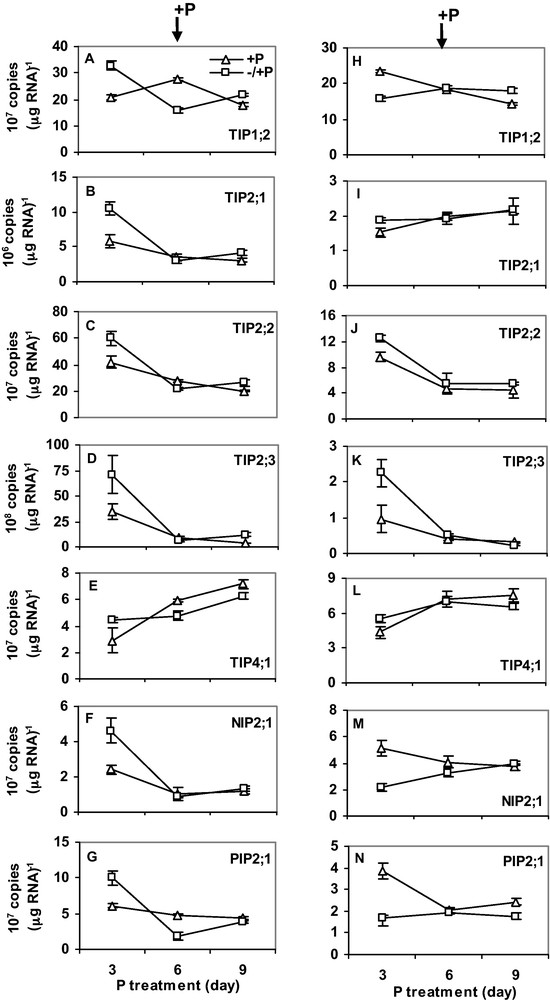
The effect of P-deficiency on the expression of MIPs in roots (A-G) and shoots (H-N). For P-treatment, P supply was resumed six days after deficiency as described in the Materials and methods. Gene expression was studied every three days. RNA was isolated from the root tips and the youngest fully expanded leaves at the same time of the day. Curves (–/ + P, open square; + P, open triangle) represent the average of two independent experiments each with four replicates, and error bars indicate SE. Note that the scale of Y-axis in the figures varies depending on the type of gene.
3.5 HvTIP2;1, HvTIP2;3 and HvNIP2;1 do not facilitate water uptake
In order to determine the physiological functions of the proteins encoded by HvTIP2;1, HvTIP2;3 and HvNIP2;1, the ORF of the genes was first cloned and their cRNA was synthesized. Oocytes were injected with the cRNA, water (negative control) or cRNA of HvPIP2;1 (positive control). Osmotic water permeability was determined by a swelling assay. Unlike oocytes injected with cRNA of HvPIP2;1, oocytes injected with HvTIP2;1, HvTIP2;3 and HvNIP2;1 did not show an increase in volume (Fig. 6). Furthermore, the osmotic water permeability (Pf, 10−2 cm s−1) of oocytes injected with cRNA of HvTIP2;1 (0.119 ± 0.015), HvTIP2;3 (0.130 ± 0.008) and HvNIP2;1(0.134 ± 0.012) was not different from oocytes injected with water (0.142 ± 0.012). While this value was 20 times higher for the positive control HvPIP2;3 (2.785 ± 0.246), indicating that the proteins encoded by HvTIP2;1, HvTIP2;3 and HvNIP2;1 do not facilitate water transport.
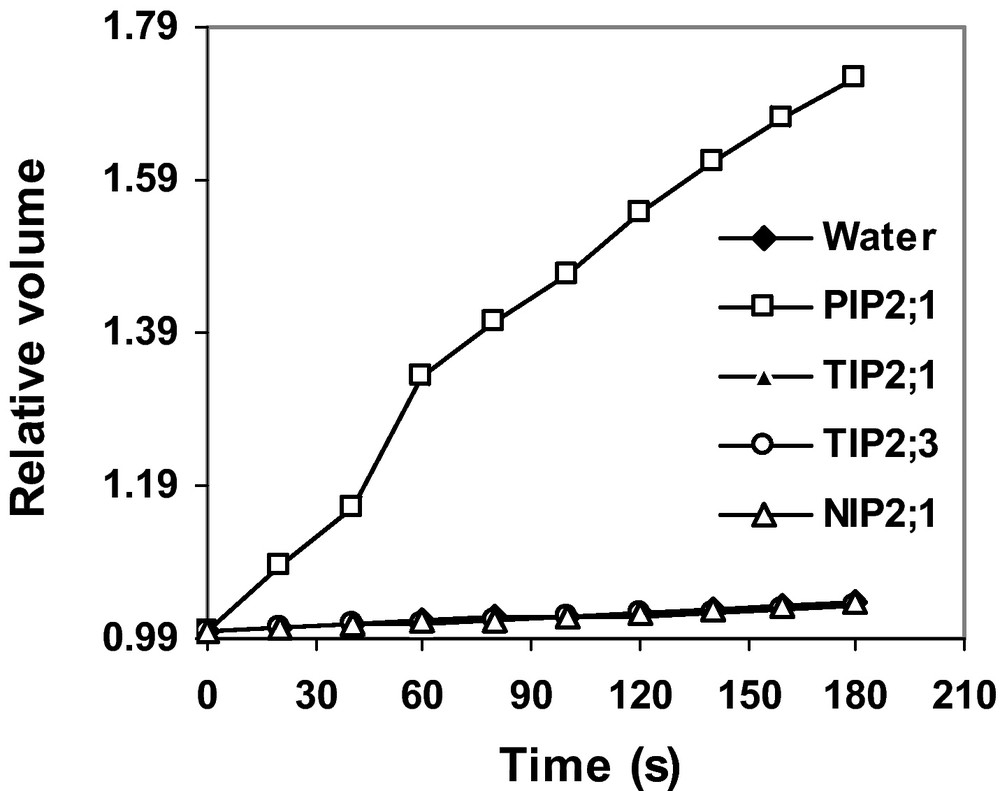
Functional analysis of HvTIP2;1, HvTIP2;3 and HvNIP2;1 proteins in X. laevis oocytes. Oocytes were injected with cRNA of HvTIP2;1, HvTIP2;3, HvNIP2;1, a positive control (HvPIP2;1) or water. Swelling assay was performed as described in the Materials and methods. Curves represent the average of two independent experiments each with 10 replicates, and error bars indicate SE.
4 Discussion
Since abiotic stresses such as salt induce osmotic stress to plants and disturb plant water balance, we tried to determine the molecular mechanisms involved in maintaining osmotic homeostasis by studying gene expression and water transport activity of MIP isoforms in barley. Transcripts of all MIPs studied were detected in both roots and shoots at different levels. Our results showed that transcript levels responded differently to 100 mM NaCl treatment (a concentration that resulted in a moderate growth reduction in barley cultivars [56] depending on the type of gene and plant tissue (Table 2). In roots, salt stress significantly upregulated transcripts of HvTIP1;2, and HvPIP2;1 but suppressed expression of HvNIP2;1 by eight-folds. Enhanced accumulation of HvTIP1;2 and HvPIP2;1 transcripts may suggest a role in salt stress tolerance. Multiple lines of evidence indicated that several MIP isoforms have been shown to enhance salt and draught stress tolerance in transgenic plants. For example, transgenic tomato plants expressing SlTIP2;2 showed improved tolerance to salt and water stresses under field conditions [57]. Similarly, a recent report from Huang's group showed that overexpression of TaNIP in transgenic Arabidopsis resulted in higher salt tolerance than wild-type plants [58]. Furthermore, overexpression of rice water channel RWC3 in upland rice enhanced root osmotic hydraulic conductivity (Lp), leaf water potential and relative cumulative transpiration suggesting its role in drought tolerance [59]. On the contrary, HvPIP2;1 which is shown to transport water when expressed in Xenopus oocytes, resulted in increased shoot/root ratio and raised salt sensitivity in transgenic rice [54,60]. Similarly, overexpression of Arabidopsis plasma membrane PIP1b aquaporin in tobacco resulted in a negative effect during drought stress, causing faster wilting [61]. These findings suggest that AQPs may mediate water loss from plant cells. In line with this, a recent finding in Katsuhara's lab showed that under hypertonic conditions induced by NaCl treatment (Horie et al., unpublished), HvPIP2 facilitate water efflux. High concentration of NaCl (200 mM) suppressed expression of PIP isoforms in barley [54], and maize [62]. Similarly, in pepper NaCl reduced PIP1 aquaporin abundance, indicating inhibitory effects of NaCl on aquaporin functionality and protein abundance [63]. Hence, the decrease in gene expression of the PIPs observed at 200 mM NaCl treatment could be an adaptation mechanism by which plants minimize water loss through efflux to survive under severe salt stress conditions. Such a high salt concentration might have also induced serious injuries to plant cells. As we reported earlier, five days of 200 mM NaCl resulted in 50 and 65% growth reduction in salt tolerant and sensitive cultivars, respectively [56]. In shoots, salt treatment enhanced expression of HvTIP2;1 and HvNIP2;1. Moreover, expression of HvNIP2;1 in shoots of three-week-old plants was significantly higher than expression in young seedlings (data not shown). Similar changes in MIPs gene expression in response to salt stress have been reported by various authors [6,33,61,64–67], and these changes seem to be tissue specific [6,31–32]. Overall, these findings indicate that expression of the MIPs is differentially regulated by salt stress.
Similar to salt stress, phytohormones differentially affected transcript accumulation. In roots, Expression of HvTIP2;1 was increased by two-folds in response to GA and ABA (Table 2), whereas in shoots, a two to three-folds increase in transcripts of HvNIP2;1 was observed while expression of HvPIP2;1 was suppressed in response to the phytohormones. Exogenous application of ABA is also reported to induce the transcript of MIPs in maize [61], Arabidopsis [33] and rice [68]. But in radish ABA and GA decreased both transcript and protein levels of MIPs belonging to PIP and TIP [67], while brassinolide did not affect gene expression. These findings suggest that expression of some MIPs involves either the ABA-dependent or ABA-independent signaling pathway based on the type of gene and plant species.
Our results showed that metals (Cd, Cu, Zn and Cr), except Hg, downregulated expression of most MIPs in roots (Table 3). Expression of HvTIP1;2, HvTIP2;2, HvNIP2,1 and HvPIP2;1 was significantly suppressed by Cd, Cu, Zn and Cr, while HvTIP4;1 was not affected by Cu, Zn and Cr. Contrary to our findings, 1 mM Cd is reported to enhance expression of HvPIP2;1 in barley root tip [69]. However, we have not observed any increase in HvPIP2;1 expression at the concentration used in the current study (200 μM). In fact, HvPIP2;1 expression was suppressed by 80%. This discrepancy could be due to a difference in experimental conditions and the barley cultivar used. Mercury, having inhibitory effect on water transport activity [17,44–45], stimulated gene expression as in the current study and as reported earlier [41]. The fact that even a brief exposure (10–25 minutes) to HgCl2 decreased Lp in Chara internodes [70] and roots of barley [45], and rice [71] indicates that a decrease in Lpr might not be due to a decrease in the abundance of water channel proteins, but may be due to a direct interaction of mercury with water channel proteins as previously shown for the mammalian aquaporin CHIP28 [72] and Arabidopsis γ-TIP [73]. Consistent with the downregulation of the MIPs in response to metals (Cd, Cu, Zn and Cr), the rate of transpiration which is greatly affected by water absorption was significantly reduced (Fig. 3). Metals except Hg decreased both transcript accumulation and rate of transpiration suggesting that these metals decrease water absorption also by suppressing the abundance of water channel proteins. Moreover, heavy metals are known to induce oxidative stress, and can result in loss of membrane integrity and inhibit the transport activity of water channels. Mercury significantly inhibited the rate of transpiration by 45%, while simultaneously enhancing transcript accumulation of the MIPs, suggesting that plants tend to overcome water shortage by compensating the water channels blocked by Hg.
Several previous reports showed that the deficiency of major nutrients such as N, P and S is reported to decrease Lpr and influence plant water status [17,50–51]. These reports prompted us to study expression of MIP genes under nutrient deficient conditions to see if the change in Lpr is transcriptionally regulated. Gene expression was markedly affected by nutrient deficiency in roots, while expression in shoots was only slightly affected (Figs. 4 and 5). Up to six days of N-deficiency suppressed transcripts of all MIPs except HvTIP4;1 more severely than control. Interestingly, transcript level was restored to control level three days after nutrient supply was resumed. Similarly, a recent report in hybrid popular showed greater transcript abundance under increased N condition [74]. Upregulation of HvTIP4;1 may suggest a role in mediating the transport of nitrogenous compounds such as NH4+, NH3 or urea as previously reported for three Arabidopsis genes (AtTIP1;2, AtTIP2;1 and AtTIP4;1) that conferred growth of urea uptake-defective yeast mutants on 2 mM urea [68]. On the other hand, transcript level was enhanced by three days of P-deficiency for all MIPs studied. However, prolonged deficiency up to six days markedly decreased gene expression. Consistent to the gene expression, previous reports have showed a decline in Lpr under nutrient deficient conditions, which was recovered to control level when deficient nutrient was supplied [17,51,71]. On the contrary, N-nutrition did not significantly affect root hydraulic conductivity in oak seedlings [75], but it was noted that nitrogen nutrition may have contributed to the reduction of Lpr of this species at higher age. This may indicate that the effect of nutrient deficiency on Lpr depends on plant species and age. In wheat roots, N and P-deficiency modulate composition and properties of plasma membrane indicating that N and P-deficiency decrease Lp by reducing either the activity or the abundance of Hg-sensitive water channel protein [17]. Inline with their suggestion, two gene transcripts in roots of L. japonicus that have a high homology to those encoding PIP1 and PIP2 AQPs of Arabidopsis found [47], and a temporal similarity between the cycles of the abundance of mRNA and Lpr has been reported. Since the plasma membrane from N and P-deficient roots of L. japonicus were not sensitive to Hg, Clarkson et al. [47] concluded that N and P-deficiency decrease Lpr by decreasing either the activity or the density of water channels in the plasma membrane. The remarkable decrease in the transcript of AQPs in the current study supports the hypothesis that N and P-deficiency modulate Lpr by decreasing the abundance of AQPs.
In order to determine the physiological roles of the MIP gene products, HvTIP2;1, HvTIP2;3 and HvNIP2;1 were expressed in Xenopus oocytes and their water transport activity was studied. Our findings revealed that oocytes injected with cRNA of HvTIP2;1, HvTIP2;3 and HvNIP2;1 did not show an increase in volume unlike oocytes injected with the cRNA of HvPIP2;1 previously reported to facilitate osmotic water uptake (Fig. 6) [54]. Moreover, the three MIPs did not increase the osmotic water permeability coefficient (Pf) compared to water injected oocytes while this value was 20 times higher for HvPIP2;1, suggesting that HvTIP2;1, HvTIP2;3 and HvNIP2;1 do not have water transport activity. The large family of MIPs (AQPs) have been known to facilitate the uptake of various molecules as mentioned in the introduction. The sequence of HvTIP2;1 and HvTIP2;3 share over 80% amino acid similarity with TIP isoforms in wheat and Arabidopsis (Fig. 2) that transport ammonia and urea, respectively [26,67], suggesting that the barley TIPs may facilitate the transport of nitrogenous compounds such as ammonia or urea. Similarly, HvNIP2;1 share over 70% similarity with NIP isoforms in maize [5], zucchini (Cucurbita pepo) [18] and rice [12] that transport glycerol, urea and Si, respectively. Therefore, it is possible that HvNIP2;1 facilitates the transport of molecules such as glycerol, urea or Si. Modulation of aquaporin activity by homomerization or hetermomerization among subunits of AQPs belonging to different subfamilies have been reported before [76–78]. In maize, coexpression of a nonfunctional ZmPIP1;1 and ZmPIP1;2 isoforms with functional ZmPIP2 protein in Xenopus oocytes significantly enhanced their aquaporin activity [77]. Further work from Chaumont's group suggested that PIP1–PIP2 interaction is required for PIP1 trafficking to the plasma membrane to modulate plasma membrane permeability [78]. Hence, MIP isoforms tested here may require heteromerization with subunits of functional AQPs to facilitate the transport of molecules.
In conclusion, we identified several MIP isoforms from barley. The Transcript of each MIP isoform is differentially regulated by various abiotic stress conditions, suggesting that the MIP isoforms have a distinct role under these stress conditions. Our findings lay a ground for future work aimed at studying the role of the MIP isoforms reported here, and other MIPs recently isolated by Katsuhara's group, in the adaptation/sensitivity to various abiotic stress conditions including salinity, drought, nutrient deficiency, heavy metals, and so on. Further research is crucial to study the subcellular localization of each MIP isoform and physiological function of MIP gene products by expressing individual MIPs or coexpressing MIPs belonging to different subgroups in Xenopus oocytes.
Conflict of interest statement
Authors declare no conflict of interest.
Acknowledgements
This research was primarily supported by a postdoctoral fellowship awarded by the Japan Society for the Promotion of Science (to A. L.); and partly by the Program for Promotion of Basic Research Activity for Innovative Bioscience (PROBRAIN, to M.K.). We thank Ms. Shizuka Sasano for technical assistance.


