1 Introduction
Cultivation of oil palm (Elaeis guineensis Jacq.) has expanded tremendously in recent years and palm oil has established itself as the leading vegetable oil in the world, taking over the top position from soybean in 2006 [1]. It is an extensive commercial crop demanding large tracts of land for its exploitation.
Knowledge on genetic diversity and genetic relationships among breeding materials could be an invaluable aid in crop improvement strategies. The use of molecular markers has provided important findings in genetic variability studies. Molecular markers can also be applied in predicting progeny performance. In a conventional breeding program, thousands of crosses were made and tested in extensive field plots. Genetic distance as measured by molecular data may be useful to predict good potential crosses, thus reducing the total number of experimental progenies.
Selection of suitable parents is one of the most important criteria used to allocate resources for the most promising crosses and increase the efficiency of breeding programs. Since hybrid vigor is contributed by genetic complementation between divergent parents, it can be assumed that parents with high genetic distance coefficients have the tendency to produce more vigorous hybrids [2]. Parents with higher general combining ability and large genetic distance produce hybrids with better yield performance [3,4]. Advances in research have generated interest in predicting progeny performance using molecular markers.
Microsatellite is one of well-known molecular markers and known as Simple Sequence Repeats (SSRs). It has short tandem repeat of DNA sequences (2–6 bp) and are highly polymorphic due to the variation in the number of repeat units. These are inherited in co-dominant fashion and highly heritable. Besides that, they are easy to score and can be produced rapidly using PCR technology. Many studies have been reported on the uses of microstellite markers. For example, microsatellites are widely used for DNA fingerprinting, linkage map construction and population genetic studies [5,6].
The objective of this study was to investigate the association between genetic distance of parental palms revealed by molecular data and performance of their Dura × Pisifera hybrids. For this purpose, simple correlation coefficient as formulated by Pearson was used [7]. We hypothesized that the information obtained from this study would help the oil palm breeders to efficiently select the promising parental palms and their progenies that have higher potential of producing more vigorous plants and omit the least potential combination for testing.
2 Materials and methods
2.1 Plant materials
A total of 19 parental palms (15 duras and four pisiferas) and 16 F1 generations from 16 crosses of Dura (D) × Pisifera (P) were employed in this study (Table 1). Each parental palm and F1 generation consisted of 19 to 20 individuals or plant samples. The Deli dura materials originated from Sabah Breeding Programme (SPB) were used as female parents. The male parents were the AVROS pisiferas, the descendents of BM119 from Oil Palm Research Station, Banting, Selangor, Malaysia. The dura palms were identified as “D” (D1–D15) whereas; pisifera palms were identified as “P” (P1–P4). The 16 F1 generations were coded as “DP” (DP1–DP16).
Number of palms parents and their F1 generations codes and their samples used in this study.
| Parents | Crosses | F1 generations | No. of Palms sample | |
| Dura Code | Pisifera Code | |||
| D1 | P1 | D1 × P1 | DP1 | 19 |
| D2 | P1 | D2 × P1 | DP2 | 20 |
| D3 | P2 | D3 × P2 | DP3 | 20 |
| D4 | P2 | D4 × P2 | DP4 | 20 |
| D5 | P2 | D5 × P2 | DP5 | 20 |
| D6 | P2 | D6 × P2 | DP6 | 20 |
| D7 | P3 | D7 × P3 | DP7 | 19 |
| D8 | P3 | D8 × P3 | DP8 | 20 |
| D2 | P3 | D2 × P3 | DP9 | 20 |
| D9 | P4 | D9 × P4 | DP10 | 20 |
| D10 | P4 | D10 × P4 | DP11 | 20 |
| D11 | P4 | D11 × P4 | DP12 | 19 |
| D12 | P1 | D12 × P1 | DP13 | 19 |
| D13 | P1 | D13 × P1 | DP14 | 19 |
| D14 | P1 | D14 × P1 | DP15 | 20 |
| D15 | P1 | D15 × P1 | DP16 | 19 |
2.2 Samples collection
Young leaves of selected parental palms were sampled from Malaysian Palm Oil Board (MPOB) Research Station, Kluang, Johore while 16 F1 (D × P) generations were collected from MPOB Research Station Keratong, Pahang. Samples were cut into small pieces, labeled and stored in liquid nitrogen during transportation to MPOB Headquarters. All the samples were stored at −80 °C until used for further analysis.
2.3 DNA extraction
DNA from oil palm leaf tissue was extracted following a modified method of Dellaporta et al. [8]. Two grams of leaf tissue were ground to fine powder in the presence of liquid nitrogen using mortar and pestle. The powder was then transferred into 30 ml Oak Ridge tube, mixed with 15 ml of extraction buffer (100 mM Tris pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl, and 10 mM merchapto-ethanol) and incubated at 60 °C for 30 mins. An equal volume of chloroform/isomyl alcohol (24:1 v/v) was then added and the mixture was centrifuged at 10,000 rpm for 20 min to separate leaf residues. The supernatant was collected into a new tube and mixed with two volume of isopropanol in order to precipitate DNA. The DNA was separated by centrifugation at 10,000 rpm for 20 mins. After discarding the supernatant, the DNA pellet was washed twice with 70% ethanol containing 10 mM ammonium acetate, dried and dissolved in TE buffer (10 mM Tris, 1 mM EDTA pH 8.0). RNase treatment was then carried out by adding RNase (50 μg/ml) and incubated at 37 °C for 20 mins. Ammonium acetate (7.5 M) and two volumes of absolute ethanol were added. The DNA was pelleted by centrifuging at 10,000 rpm for 20 mins. After washing with 70% ethanol, the pellet was dissolved in 1 ml of TE buffer.
2.4 Digestibility test and quantification of oil palm DNA
Two restriction enzymes, EcoR1 and HaeIII were used for digestibility test of DNA. Both digested and undigested DNA was loaded into 0.8% agarose gel and electrophoresis was done at 100 V in 1X TAE buffer. The gel was stained in ethidium bromide and visualized under UV light. The image was captured using a Polaroid camera. DNA concentration was determined using spectrophotometer. Optical density (OD) was noted at wavelength 260 and 280 nm. The concentration of DNA was calculated from OD reading at 260 nm while DNA purity was calculated by the ratio of absorbance obtained at 260 and 280 nm. The ratio of good DNA quality ranged from 1.8 to 2.2.
2.5 Microsatellite markers
A sum of nine microsatellite primers, out of 12 primers tested earlier, were developed in MPOB and derived from expressed sequence tag database [9]. These primers were chosen based on their ability to generate polymorphic and scorable amplification products. The annealing temperature of the primers ranged from 52 to 54 °C (Table 2).
List of the primers used in microsatellite analyses.
| No. | Primer | TA (°C) | 5’ sequence | 3’ sequence | Allele size (bp) | Repeat unit |
| 1 | CNH00887 | 52 °C | TTATTGATTGATGCAAGATACAC | TTGATAAAATACAAGAGATAGCA | 165 | (AT)9 |
| 2 | CNH00938 | 52 °C | GGACCCTTTTTGTTACTGTTT | AGCCTACCACAACTTCCTTT | 172 | (AG)9 |
| 3 | CNH01617 | 52 °C | TCTTTAATTTGTCGAGGATAATG | ATGCAAGGTTTTGTTGAAACT | 130 | (CT)20 |
| 4 | CNI01937 | 52 °C | AACTGCAAATGAGACACAGAG | TCCACCAGAGGAGGGTTAGT | 170 | (AG)9 |
| 5 | EAP 03160 | 52 °C | AACGTGAGAGCCATAGAGATAG | TAATAGAAACTAGACCCGACCA | 175 | (TATG)6 |
| 6 | EO 02978 | 52 °C | CCGTCTCAAAAGCCCTAAAC | TTGTTGTCCCACTCCCTCTT | 210 | (CGC)7 |
| 7 | MF233033 | 52 °C | GAGGAGGAGGGGAGAAGAGT | AAATACCATTCAGAGAAAGCAC | 200 | (TC)11 |
| 8 | MF233056 | 52 °C | CCGAATAGAAGAGGAAAGAATA | AGGTTTGGTGGAGAAGTGTT | 232 | (CT)15 |
| 9 | MF2331019 | 54 °C | TGGGTAAATTGGTAATTCTCCT | CCTTTTTCTTCCTCTTTTCCA | 195 | (TC)8 |
2.6 PCR protocols
PCR reaction mixture contained primer mix (T4 polynucleotide kinase, (−33P-ATP, microsatellite primers and kinase buffer), 5U/μl Taq DNA polymerase, 10 mM dNTPs, 50 mM MgCl2, 10X PCR buffer and 1 μl of template DNA. DNA template was diluted to final concentration of 50 ng/μl. PCR was performed in a thermocycler (Perkin Elmer 9600) essentially as described by Billotte et al. [10]. The PCR amplification was done as follows: denaturation at 95 °C for 30 seconds, annealing at 52 to 54 °C (depending on the primer used) for 30 seconds, extension at 72 °C for 30 seconds; all of these steps were repeated for 35 cycles.
2.7 Polyacrylamide gel electrophoresis (PAGE) and gel scoring
Mixture of final PCR product, bromophenol blue and xylene cyanol were denatured at 90 °C. Bromophenol blue and xylene cyanol were used to show migration because they give color to the PCR product. The samples were run in 6% PAGE at 1600 V for 2 to 3 h. The polyacrylamide gels (30 cm × 30 cm) were then vacuumed dried for an hour and exposed against X-ray film (Kodak) for 3 to 4 days at –80 °C depending on the radioactive signal on the gel. Microsatellite banding patterns observed in 16 F1 generations and their parental palms were transformed into alleles and loci. Each band within each locus was identified according to alphabetical order and beginning with the most anodal migrating band designated as allele A, the next band was allele B or otherwise (Table 3). With microsatellites, one can usually identify more than two alleles per locus.
Number of alleles and assignment of alleles for each primer used in molecular study.
| No. | Primer | No. of allele produced | Assignment of alleles | |
| 1 | CNH00887 | 4 | A, B, C, D | 106 |
| 2 | CNH00938 | 3 | A, B, C | 107 |
| 3 | CNH01617 | 3 | A, B, C | 108 |
| 4 | CNI 01937 | 2 | A, B | 109 |
| 5 | EAP 03160 | 2 | A, B | 110 |
| 6 | EO 02978 | 2 | A, B | 111 |
| 7 | MF233033 | 4 | A, B, C, D | 112 |
| 8 | MF233056 | 6 | A, B, C, D, E, F | 113 |
| 9 | MF2331019 | 3 | A, B, C | |
| Total | 29 | |||
| Mean | 3.22 |
2.8 Field evaluation of the F1 (D × P) generations
For field performances, 16 F1 generations were evaluated. In each generation, a total of 19 to 20 individuals were used in measurement of 29 quantitative characters such as yield, yield components and vegetative characters (Appendix 1). Yield data were collected for 7 years. Oil palm usually starts fruiting about 27 months after field planting. The first fruit normally ripen during 3rd year after field planting. Thus, yield recording only commenced at 36 months [11]. Each palm was inspected after every 10 days and any ripen fruit bunch present was harvested and weighed using a simple spring balance. The weight and number of bunches were recorded for each palm. The vegetative measurement is the measure of physiological and vegetative performance of a palm. The vegetative data were scored using non-destructive method proposed by Corley and Breure [12].
2.9 Data analysis
For molecular analysis, scorable fragments were transformed to genotypic data and arranged as a data matrix. Data were analyzed using Biosys1 computer program to generate distance coefficients. Genetic distances among populations were computed according to the method described by Rogers [13].
Mean data of 29 characters of 16 F1 generations, obtained during field evaluation, were analyzed by principle coordinate analysis using GENSTAT 5.13 software program (copyright 1987, Lawes agricultural Trust, Rothamasted Experimental station, UK). Estimation of distance between the generations was determined and a dendrogram was constructed using distance matrix. Correlation analyses were performed using Pearson's coefficient to determine the correlation level among the microsatellite-based genetic distance and hybrid performance [7] using the SAS program.
3 Results
3.1 Microsatellites polymorphisms
Nine microsatellite primers showed 29 polymorphic bands in parental palms and their F1 generations. In general, two to six bands were scored per primer (Table 3). Primer MF233056 produced the highest alleles (6) compared to other primers. Four alleles were produced by primer CNH 00887 and MF233033. Primer CNH 00938, CNH 01617 and MF2331019 produced three alleles. Primer CNI 01937, EAP 03160 and EO 02978 produced only two alleles (Fig. 1 and Table 3).
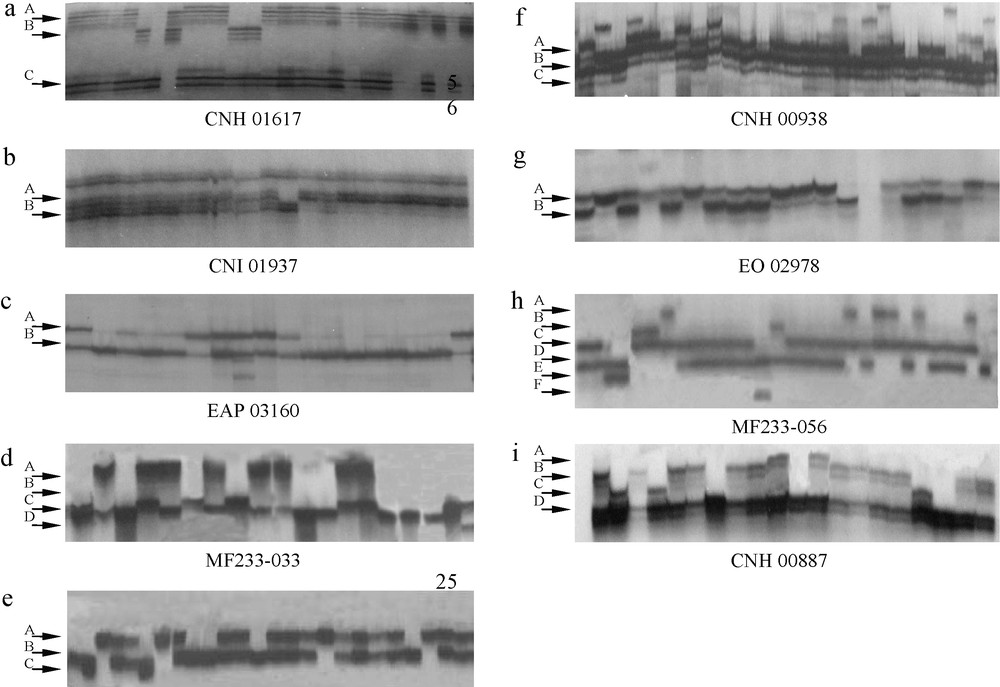
Autoradiogram amplified from nine different microsatellite primers showing profile of F1 progenies (A to F refer to allele types; A: Allele A; B: Allele B; C: Allele C; D: Allele D; E: Allele E; F: Allele F).
In the case of parental lines, genetic variability was calculated and presented in Table 4. The mean number of alleles per locus ranged from 2.0 to 2.2, with a grand mean of 2.1. The percentage of polymorphic loci at 0.95 criterion ranged from 88.5 to 100% with average of 97.0%.
Genetic variability of 19 parental oil palms.
| Population (Parents) | Mean sample size per locus | Mean no. of allele per locus | Percentage of polymorphic locia | Mean Heterozygosity | |
| Observed (Ho) | Expected (He) | ||||
| 1. D1 | 19.0 | 2.2 | 98.0 | 0.587 | 0.448 |
| 2. D2 | 18.0 | 2.1 | 98.0 | 0.557 | 0.432 |
| 3. D3 | 19.0 | 2.2 | 98.0 | 0.562 | 0.414 |
| 4. D4 | 18.0 | 2.1 | 100.0 | 0.654 | 0.460 |
| 5. D5 | 20.0 | 2.2 | 100.0 | 0.653 | 0.478 |
| 6. D6 | 19.0 | 2.3 | 100.0 | 0.653 | 0.498 |
| 7. D7 | 18.3 | 2.2 | 100.0 | 0.685 | 0.477 |
| 8. D8 | 20.0 | 2.0 | 98.9 | 0.596 | 0.437 |
| 9. D9 | 20.0 | 2.1 | 98.9 | 0.598 | 0.440 |
| 10. D10 | 18.0 | 2.2 | 98.9 | 0.595 | 0.439 |
| 11. D11 | 18.4 | 2.1 | 98.9 | 0.599 | 0.442 |
| 12. D12 | 18.5 | 2.2 | 100.0 | 0.661 | 0.494 |
| 13. D13 | 19.0 | 2.0 | 100.0 | 0.607 | 0.490 |
| 14. D14 | 18.8 | 2.2 | 100.0 | 0.634 | 0.492 |
| 15. D15 | 19.0 | 2.0 | 100.0 | 0.696 | 0.495 |
| 16. P1 | 18.4 | 2.1 | 88.5 | 0.583 | 0.471 |
| 17. P2 | 18.0 | 2.1 | 88.5 | 0.583 | 0.471 |
| 18. P3 | 18.4 | 2.1 | 88.5 | 0.580 | 0.450 |
| 19. P4 | 18.4 | 2.1 | 88.5 | 0.580 | 0.450 |
| Mean | 18.7 | 2.1 | 97.0 | 0.613 | 0.462 |
a A locus is considered polymorphic if the frequency of the most common allele does not exceed 0.95.
The grand means observed (Ho) and expected (He) heterozygosities across populations were 0.613 and 0.462, respectively. The mean expected heterozygosity values ranged from 0.414 (D3) to 0.498 (D6). The mean observed heterozygosity was the lowest for D2 (0.557) and the highest for D15 (0.696).
Genetic variability of F1 generations are presented in Table 5. The mean number of alleles per locus ranged from 1.9 to 2.6, with an average of 2.3. The percentage of polymorphic loci at 0.95 criterion ranged from 88.9 to 100% with an average of 94.5%. The mean observed (Ho) and expected (He) heterozygosities across populations were found to be 0.621 and 0.455, respectively. The mean expected heterozygosity values ranged from 0.387 (DP8) to 0.498 (DP6 and DP1). The mean observed heterozygosity was the lowest for DP3 (0.512) and the highest for DP11 (0.722).
Genetic variability of 16 F1 (D × P) generations of oil palm.
| Population F1 generations) | Mean sample size per locus | Mean no. of allele per locus | Percentage of polymorphic locia | Mean Heterozygosity | |
| Observed (Ho) | Expected (He) | ||||
| 1. DP1 | 18.8 | 2.4 | 100.0 | 0.686 | 0.498 |
| 2. DP2 | 20.0 | 2.4 | 100.0 | 0.656 | 0.492 |
| 3. DP3 | 19.7 | 2.4 | 100.0 | 0.512 | 0.414 |
| 4. DP4 | 19.0 | 2.1 | 88.9 | 0.556 | 0.433 |
| 5. DP5 | 20.0 | 2.1 | 88.9 | 0.544 | 0.389 |
| 6. DP6 | 20.0 | 2.3 | 100.0 | 0.683 | 0.498 |
| 7. DP7 | 18.3 | 2.1 | 100.0 | 0.588 | 0.427 |
| 8. DP8 | 20.0 | 1.9 | 88.9 | 0.556 | 0.387 |
| 9. DP9 | 20.0 | 2.1 | 88.9 | 0.628 | 0.435 |
| 10. DP10 | 18.8 | 2.4 | 88.9 | 0.605 | 0.491 |
| 11. DP11 | 18.0 | 2.3 | 88.9 | 0.722 | 0.482 |
| 12. DP12 | 19.7 | 2.6 | 100.0 | 0.671 | 0.484 |
| 13. DP13 | 18.4 | 2.2 | 88.9 | 0.607 | 0.469 |
| 14. DP14 | 18.8 | 2.4 | 100.0 | 0.634 | 0.491 |
| 15. DP15 | 19.3 | 2.4 | 100.0 | 0.596 | 0.417 |
| 16. DP16 | 18.4 | 2.2 | 88.9 | 0.685 | 0.471 |
| Mean | 19.0 | 2.3 | 94.5 | 0.621 | 0.455 |
a A locus is considered polymorphic if the frequency of the most common allele does not exceed 0.95.
3.2 Genetic distance-based analysis for 19 Parental Palms using molecular markers
Table 6 showed the values of genetic distance among the dura and pisifera parental palms. Cluster analysis indicated a clear separation of dura and pisifera parental palms (Fig. 2). The Rogers's distance values for dura parental palms ranged from 0.050 to 0.573. The lowest genetic distance corresponded to dura parental palms, D9 and D10 with value of 0.05. On the other hand, the highest genetic distance was found between palms, D2 and D14 with a value of 0.573. A value of 0.573 indicated that there were 57.3% dissimilarities among the populations in the portion of genome surveyed by the nine microsatellite markers. The genetic distance between dura and pisifera parental palms showed that the highest genetic distance was found between palms, D11 and P4 (0.746). On the other hand, the lowest genetic distance was detected between palms, D2 and P1 (0.444) (Table 6).
Genetic distance of 19 parental palms based on nine microsatellite markers.
| Parental lines | D2 | D4 | D3 | D5 | D11 | D8 | D13 | D6 | D14 | D1 | D12 | D7 | D9 | D10 | D15 | P1 | P3 | P2 | P4 |
| D2 | ***** | ||||||||||||||||||
| D4 | 0.371 | ***** | |||||||||||||||||
| D3 | 0.250 | 0.400 | ***** | ||||||||||||||||
| D5 | 0.221 | 0.250 | 0.200 | ***** | |||||||||||||||
| D11 | 0.250 | 0.400 | 0.437 | 0.387 | ***** | ||||||||||||||
| D8 | 0.321 | 0.250 | 0.300 | 0.250 | 0.287 | ***** | |||||||||||||
| D13 | 0.337 | 0.400 | 0.187 | 0.337 | 0.487 | 0.400 | ***** | ||||||||||||
| D6 | 0.300 | 0.437 | 0.400 | 0.450 | 0.400 | 0.450 | 0.423 | ***** | |||||||||||
| D14 | 0.573 | 0.437 | 0.500 | 0.500 | 0.550 | 0.450 | 0.537 | 0.400 | ***** | ||||||||||
| D1 | 0.350 | 0.300 | 0.300 | 0.321 | 0.521 | 0.450 | 0.250 | 0.478 | 0.473 | ***** | |||||||||
| D12 | 0.43 | 0.407 | 0.521 | 0.350 | 0.450 | 0.371 | 0.487 | 0.521 | 0.323 | 0.421 | ***** | ||||||||
| D7 | 0.237 | 0.350 | 0.300 | 0.150 | 0.387 | 0.350 | 0.437 | 0.400 | 0.450 | 0.423 | 0.473 | ***** | |||||||
| D9 | 0.300 | 0.350 | 0.300 | 0.300 | 0.350 | 0.200 | 0.373 | 0.387 | 0.300 | 0.387 | 0.307 | 0.300 | ***** | ||||||
| D10 | 0.250 | 0.350 | 0.250 | 0.250 | 0.300 | 0.250 | 0.323 | 0.437 | 0.350 | 0.337 | 0.357 | 0.250 | 0.050 | ***** | |||||
| D15 | 0.250 | 0.437 | 0.300 | 0.337 | 0.487 | 0.337 | 0.387 | 0.337 | 0.437 | 0.437 | 0.460 | 0.350 | 0.200 | 0.250 | ***** | ||||
| P1 | 0.444 | 0.387 | 0.528 | 0.437 | 0.494 | 0.423 | 0.507 | 0.515 | 0.628 | 0.557 | 0.537 | 0.473 | 0.494 | 0.544 | 0.473 | ***** | |||
| P3 | 0.687 | 0.473 | 0.587 | 0.587 | 0.737 | 0.587 | 0.550 | 0.660 | 0.687 | 0.473 | 0.637 | 0.737 | 0.637 | 0.687 | 0.600 | 0.400 | ***** | ||
| P2 | 0.573 | 0.460 | 0.687 | 0.587 | 0.587 | 0.587 | 0.650 | 0.507 | 0.473 | 0.523 | 0.387 | 0.623 | 0.623 | 0.673 | 0.623 | 0.400 | 0.400 | ***** | |
| P4 | 0.710 | 0.481 | 0.644 | 0.607 | 0.746 | 0.596 | 0.607 | 0.651 | 0.565 | 0.494 | 0.494 | 0.744 | 0.633 | 0.683 | 0.623 | 0.350 | 0.300 | 0.300 | ***** |
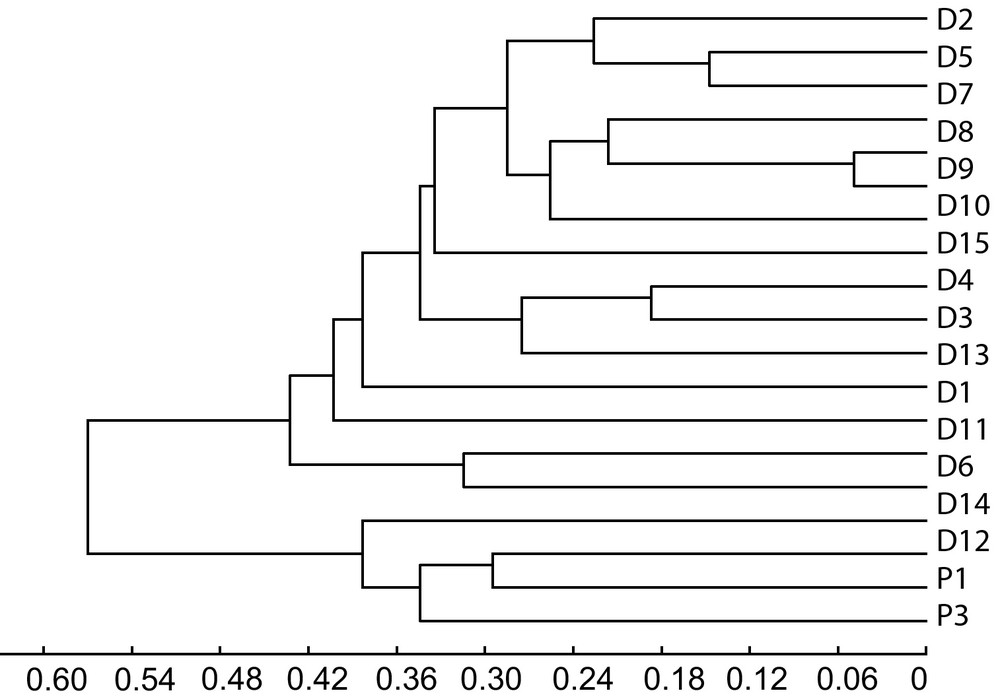
Dendrogram showing UPGMA clusters for 19 parental palms based on molecular markers.
3.3 Genetic distance-based analysis for 16 F1 (D × P) generations using molecular markers
The genetic distance between 16 F1 (D × P) generations is presented in Table 7. The values ranged from 0.089 (between DP10 and DP11) to 0.313 (between DP8 and DP16). There was no 0 (zero) value indicating that no population was identical. From the pedigree information, DP10 and DP11 had common male parental palms (P4). Besides that, the female parental palms of these progenies were found to be the siblings. DP8 and DP16 had the highest genetic distance value.
Genetic distance of 16 F1 generations based on nine microsatellite markers.
| Progeny | DP2 | DP9 | DP4 | DP3 | DP5 | DP12 | DP8 | DP14 | DP6 | DP15 | DP1 | DP13 | DP7 | DP10 | DP11 | DP16 |
| DP2 | **** | |||||||||||||||
| DP9 | 0.179 | **** | ||||||||||||||
| DP4 | 0.307 | 0.300 | **** | |||||||||||||
| DP3 | 0.265 | 0.284 | 0.295 | **** | ||||||||||||
| DP5 | 0.292 | 0.295 | 0.209 | 0.189 | **** | |||||||||||
| DP12 | 0.163 | 0.193 | 0.280 | 0.139 | 0.196 | **** | ||||||||||
| DP8 | 0.265 | 0.228 | 0.244 | 0.174 | 0.132 | 0.197 | **** | |||||||||
| DP14 | 0.105 | 0.208 | 0.243 | 0.191 | 0.209 | 0.107 | 0.210 | **** | ||||||||
| DP6 | 0.235 | 0.264 | 0.234 | 0.218 | 0.234 | 0.203 | 0.252 | 0.203 | **** | |||||||
| DP15 | 0.267 | 0.261 | 0.234 | 0.135 | 0.111 | 0.163 | 0.120 | 0.192 | 0.192 | **** | ||||||
| DP1 | 0.189 | 0.214 | 0.235 | 0.203 | 0.197 | 0.176 | 0.161 | 0.168 | 0.228 | 0.137 | **** | |||||
| DP13 | 0.157 | 0.242 | 0.265 | 0.193 | 0.185 | 0.091 | 0.226 | 0.090 | 0.227 | 0.188 | 0.196 | **** | ||||
| DP7 | 0.163 | 0.178 | 0.296 | 0.258 | 0.223 | 0.193 | 0.214 | 0.199 | 0.307 | 0.205 | 0.173 | 0.209 | **** | |||
| DP10 | 0.198 | 0.229 | 0.247 | 0.209 | 0.174 | 0.114 | 0.147 | 0.137 | 0.226 | 0.144 | 0.177 | 0.126 | 0.210 | **** | ||
| DP11 | 0.165 | 0.230 | 0.271 | 0.253 | 0.193 | 0.142 | 0.200 | 0.147 | 0.247 | 0.173 | 0.178 | 0.161 | 0.186 | 0.089 | **** | |
| DP16 | 0.100 | 0.212 | 0.299 | 0.278 | 0.307 | 0.182 | 0.313 | 0.137 | 0.221 | 0.276 | 0.235 | 0.161 | 0.205 | 0.221 | 0.175 | **** |
The results of cluster analysis in the form of dendrogram are presented in Fig. 3. Four clusters were formed. Two clusters had single population (DP4 and DP6), which were separated from other clusters. The first cluster was divided into 3 sub-clusters. Sub-cluster 1 contained DP2 and DP16, which shared one male parent, P1. Sub-cluster 2 consisted of DP12, DP13 and DP14. DP13 and DP14 had a common male parental palm, P1. Meanwhile, in sub-cluster 3, DP9 and DP7 had a same male parent, P3. The second cluster consisted of DP3, DP5, DP15, DP8 and DP1. These DPs were inter-related because they were produced from the same male and female grandparents. In addition, DP3 and DP5 had common pisifera parent where as DP8 and DP1 had come from the same male grandparent.
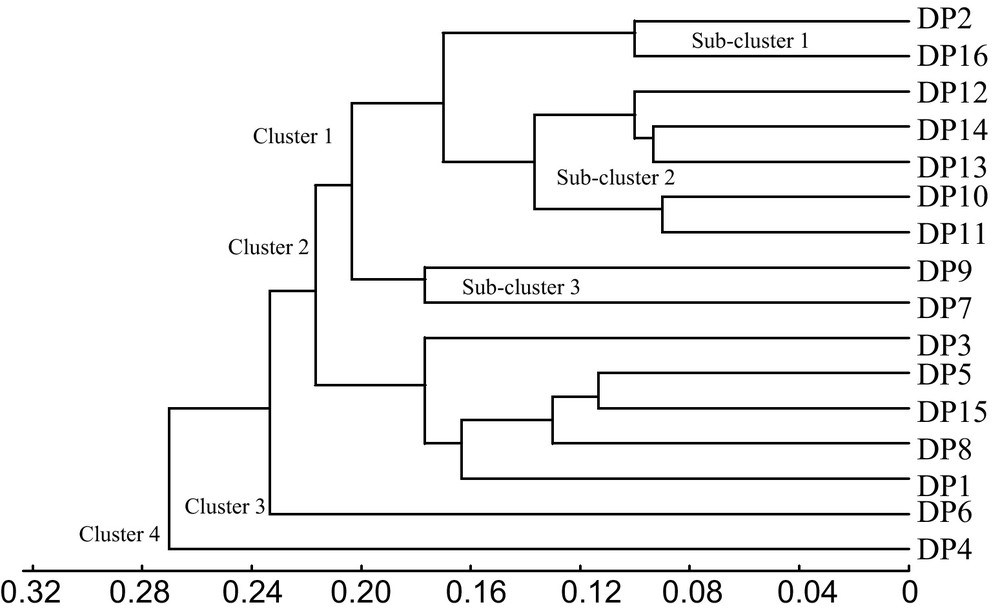
Dendrogram showing UPGMA clusters for 16 F1 generations based on molecular markers.
3.4 Genetic distance-based analysis for 16 F1 (D × P) generations using quantitative characters
Quantitative genetic distance between different pairs of different progenies was calculated. The pair-wise genetic distance indicated that the highest genetic distance was obtained between DP16 and DP14 (2.2026) (Table 8) followed by DP4 and DP14 (2.1809), DP16 and DP13 (2.0974), DP4 and DP12 (2.0846), DP4 and DP15 (2.0700), DP4 and DP8 (2.0381), DP4 and DP1 (2.0330) and DP4 and DP2 (2.0175).
Genetic distance of 16 F1 generations based on 29 quantitative characters.
| DP1 | DP2 | DP3 | DP4 | DP5 | DP6 | DP7 | DP8 | DP9 | DP10 | DP11 | DP12 | DP13 | DP14 | DP15 | DP16 | |
| DP1 | 0.0000 | |||||||||||||||
| DP2 | 1.1315 | 0.0000 | ||||||||||||||
| DP3 | 1.2602 | 1.2052 | 0.0000 | |||||||||||||
| DP4 | 2.0330 | 2.0175 | 1.4552 | 0.0000 | ||||||||||||
| DP5 | 1.5456 | 1.6571 | 1.0088 | 1.4440 | 0.0000 | |||||||||||
| DP6 | 0.8813 | 1.1747 | 1.0646 | 1.7665 | 1.4105 | 0.0000 | ||||||||||
| DP7 | 1.5917 | 1.3739 | 1.3124 | 1.6120 | 1.2104 | 1.6761 | 0.0000 | |||||||||
| DP8 | 1.0197 | 1.0089 | 0.9605 | 2.0381 | 1.3336 | 1.0661 | 1.3100 | 0.0000 | ||||||||
| DP9 | 1.4208 | 1.1739 | 1.3524 | 1.9412 | 1.4459 | 1.6479 | 0.7934 | 1.1485 | 0.0000 | |||||||
| DP10 | 0.9095 | 1.2755 | 1.1012 | 1.7190 | 1.1366 | 1.2483 | 1.0950 | 0.9484 | 0.9728 | 0.0000 | ||||||
| DP11 | 0.7531 | 1.1041 | 1.3616 | 1.9286 | 1.4405 | 1.1797 | 1.2828 | 1.1482 | 1.3390 | 0.8925 | 0.0000 | |||||
| DP12 | 0.9293 | 0.8951 | 1.2018 | 2.0846 | 1.4838 | 1.2427 | 1.2712 | 0.9128 | 1.2829 | 1.0834 | 0.8335 | 0.0000 | ||||
| DP13 | 1.3888 | 1.2567 | 1.5444 | 1.8789 | 1.6702 | 1.5105 | 1.2062 | 1.5557 | 1.4966 | 1.3102 | 1.0540 | 0.9861 | 0.0000 | |||
| DP14 | 1.1919 | 1.2241 | 1.7180 | 2.1809 | 1.9127 | 1.4157 | 1.6183 | 1.5887 | 1.5329 | 1.4135 | 1.2528 | 1.1231 | 0.9804 | 0.0000 | ||
| DP15 | 0.7864 | 1.0956 | 1.3641 | 2.0700 | 1.6742 | 1.0746 | 1.5395 | 1.1157 | 1.4144 | 1.1227 | 1.0819 | 0.8388 | 1.1744 | 0.7112 | 0.0000 | |
| DP16 | 1.9378 | 1.6774 | 1.2328 | 1.4568 | 1.4714 | 1.7890 | 1.5601 | 1.5818 | 1.4958 | 1.5323 | 1.9635 | 1.9243 | 2.0974 | 2.2026 | 1.9195 | 0.0000 |
The genetic distance analysis using UPGMA clustering system generated 5 genetic clusters with correlation coefficient of 1.26. Here, DP4 and DP16 formed 2 separate clusters. The progenies, DP7 and DP9 grouped together while DP3 and DP5 produced another cluster (Fig. 4). Ten progenies of oil palm (DP15, DP14, DP13, DP6, DP2, DP10, DP8, DP, DP12, DP11 and DP1) were clustered distinctly in the same group.
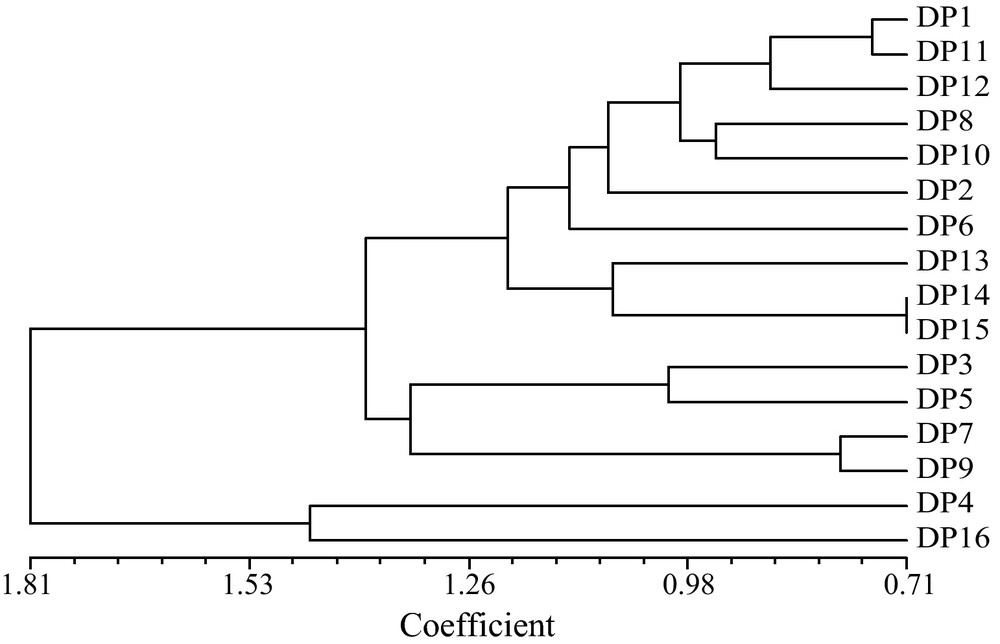
Dendrogram showing UPGMA clusters for 16 F1 generations based on 29 phenotypic characters.
3.5 Relation of progeny performance with molecular markers based genetic distances
The results of correlation analyses showed that there was no relationship between genetic distance of parental palms and the performance of the progenies for yield components and vegetative characters except mean nut weight and leaflet number (Table 9). At p ≤ 0.05, significant negative correlation was found for mean nut weight (−0.51) and leaflet number (−0.55).
Correlation between quantitative characters measured on 16 F1 generations and genetic distance of the parents.
| Yield component (n = 16) | Coefficient of correlation (r) |
| Fresh fruit bunch | 0.37 ns |
| Bunch number | 0.48 ns |
| Average Bunch weight | 0.00 ns |
| Mean fruit weight | −0.45 ns |
| Mean nut weight | −0.51a |
| Mesocarp to fruit ratio | 0.24 ns |
| Kernel to fruit ratio | −0.18 ns |
| Shell to fruit ratio | −0.26 ns |
| Oil to dry mesocarp ratio | −0.19 ns |
| Oil to wet mesocarp ratio | −0.04 ns |
| Fruit to bunch ratio | 0.01 ns |
| Oil to bunch ratio | 0.12 ns |
| Kernel to bunch ratio | −0.14 ns |
| Oil yield | 0.39 ns |
| Kernel yield | 0.11 ns |
| Total oil | 0.36 ns |
| Total economic product | 0.35 ns |
| Frond production | 0.21 ns |
| Petiole cross-section | −0.06 ns |
| Rachis length | 0.06 ns |
| Leaflet length | −0.18 ns |
| Leaflet width | 0.32 ns |
| Leaflet number | −0.55a |
| Palm height | 0.39 ns |
| Leaflet area | −0.02 ns |
| Trunk diameter | −0.50 ns |
| Leaflet area index | −0.02 ns |
| Frond index | 0.06 ns |
| Leaf area ratio | 0.07 ns |
a Significant at p ≤ 0.05.
4 Discussion
In microsatellite primer, the mean number of alleles per population of parental palms and progenies was quite high compared to other oil palm natural populations using isozyme with a mean of 1.6 [14] and RFLP technique with a mean of 1.8 [15]. However, the value was lower than that of reported for monocot species such as coconut (4.83) [16]. For progenies, the percentage of polymorphic loci at 0.95 criterions ranged from 88.9 to 100% with an average of 94.5%. Milbourne et al. [17] found that microsatellites are the better method consistently detected the highest level of polymorphism in barley (100%) and potato (90.8%). The mean observed (Ho) and expected (He) heterozygosities across populations were 0.621 and 0.455, respectively. The expected heterozygosity obtained in the palm progenies was high when compared to other monocot species such as banana (He = 0.411) [18] and Antirhea aromatica (He = 0.185) [19]. The high value of observed heterozygosity in the oil palm populations screened in this study might have resulted from the impact of selection, which generally favors heterozygote genotypes. In general, selection pressure does not cause inbreeding. In spite of extensive selection imposed on the dura and pisifera parental palms, the progenies retained considerably high levels of genetic variation. Cluster analysis indicated a clear separation of dura and pisifera parental palms. The highest genetic distance was found between palms, D11 and P4 with a value of 0.746. According to Corley et al. [20], oil palm parents with greater genetic distance generate more variable offsprings. They reported that genetic distance can be predicted from the limited ancestral information within breeding program.
For progenies performances, the genotypes, DP16, DP14, DP4 DP13, DP12, DP15, DP4, DP8, DP1 and DP2 belonging to the distant clusters and higher genetic distances could be selected for further breeding program. Similar findings of genetic divergence are also reported by several authors [21–24].
The genetic distances among parental palms were not related with the performance of their progenies. Similar results were obtained by Martin et al. [25] and Barbosa-Neto et al. [26] in their studies on wheat. They reported non-significant relationship between marker-based genetic distance and hybrid performance. Results reported by Cerna et al. [27] in soybean and Diers et al. [4] in oilseed rape also showed that correlations were not significant and not applicable for prediction of performance. Significantly (p ≤ 0.05) negative correlation was found for mean nut weight (−0.51) and leaflet number (LN) (−0.55). From the results, it is expected that genetic distance with higher value would have the lower production of leaves. Even though leaflet number was significantly negatively correlated with genetic distance, the value was too low to be predictive in estimating progeny performance. As reported by Melchinger et al. [28], inadequate genome coverage and different levels of dominance among hybrids could result in the observed low correlation between genetic distance and hybrid performance. On the contrary, Arcade et al. [29] found significant correlations between parental genetic distance and hybrid performance in larch. Besides that, Smith et al. [30] observed a significant relationship between parental genetic distance and F1 performance when the sample size as well as the number of markers used for analysis was increased simultaneously. Previous studies on predicting progeny performance using genetic markers reported similar results. Godshalk et al. [31] and Dudley et al. [32] observed weak correlations between marker genotype and hybrid performance in maize. Martin et al. [25] found no association between measures of diversity and hybrid performance in wheat (Triticum aestivum L.).
5 Conclusion
On the basis of the results it may be concluded that the correlation coefficient of genetic distances with hybrid performance were non-significant, except for mean nut weight and leaf number. The correlation of genetic distances with 29 quantitative traits was low to be used as predicted value. So, the genetic distances based on the microsatellite markers may not be useful for predicting hybrid performance. UPGMA clustering system generated five genetic clusters based on 29 quantitative characters. To exploit heterosis genotypes, DP16, DP14, DP4, DP13, DP12, DP15, DP8, DP1 and DP2 belonging to the distant clusters and greater genetic distances could be selected for hybridization program.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgement
The authors are grateful to Malaysian Palm Oil Board (MPOB) and Universiti Putra Malaysia for supporting this research project.
Appendix A Mean data of 29 quantitative characters of 16 F1 generations of oil palm
| Progenies | MFW (g) | MNW (g) | M/F (%) | K/F (%) | S/F (%) | O/DM (%) | O/WM (%) | F/B (%) | O/B (%) | K/B (%) | OY (kg) | KY (kg) | TOIL (kg p−1 yr−1) | TEP (kg p−1 yr−1) | |
| DP1 | 10.28 | 2.15 | 78.94 | 9.54 | 11.51 | 78.61 | 48.44 | 66.68 | 25.52 | 6.38 | 33.32 | 8.4 | 37.52 | 38.35 | |
| DP2 | 9.78 | 1.89 | 80.30 | 9.39 | 10.31 | 79.40 | 47.75 | 62.31 | 24.07 | 5.91 | 34.82 | 8.84 | 39.24 | 40.13 | |
| DP3 | 9.54 | 1.79 | 78.78 | 9.08 | 11.14 | 79.12 | 50.71 | 62.99 | 25.12 | 5.95 | 27.38 | 6.43 | 30.6 | 31.24 | |
| DP4 | 11.63 | 2.69 | 76.53 | 9.15 | 14.32 | 78.79 | 48.79 | 62.69 | 23.18 | 5.89 | 20.68 | 5.57 | 23.46 | 24.02 | |
| DP5 | 9.47 | 1.88 | 79.41 | 7.98 | 12.61 | 79.02 | 48.63 | 69.03 | 26.58 | 5.52 | 28.73 | 6 | 31.37 | 32.32 | |
| DP6 | 11.01 | 2.45 | 77.38 | 9.53 | 13.10 | 80.21 | 51.06 | 65.39 | 25.9 | 6.18 | 32.31 | 7.51 | 36.07 | 36.82 | |
| DP7 | 10.01 | 1.77 | 81.64 | 8.09 | 10.27 | 78.24 | 47.58 | 61.97 | 24.12 | 4.99 | 33.18 | 6.79 | 36.58 | 37.25 | |
| DP8 | 8.57 | 1.54 | 81.59 | 8.91 | 9.50 | 79.85 | 50.60 | 61.87 | 25.58 | 5.5 | 35.22 | 7.37 | 38.9 | 39.64 | |
| DP9 | 10.21 | 1.49 | 84.70 | 7.78 | 7.52 | 78.31 | 46.63 | 61.77 | 24.17 | 4.89 | 32.38 | 6.4 | 35.58 | 36.21 | |
| DP10 | 9.29 | 1.77 | 80.59 | 8.58 | 10.83 | 77.67 | 47.17 | 65.02 | 24.59 | 5.59 | 31.84 | 7.12 | 35.4 | 36.11 | |
| DP11 | 10.28 | 2.19 | 78.18 | 9.90 | 11.92 | 78.18 | 47.24 | 65.91 | 24.42 | 6.48 | 35.77 | 9.44 | 40.49 | 41.43 | |
| DP12 | 7.72 | 1.65 | 78.65 | 9.83 | 11.52 | 78.65 | 49.15 | 64.76 | 24.95 | 6.39 | 35.31 | 9.1 | 39.86 | 40.77 | |
| DP13 | 8.72 | 2.04 | 76.24 | 9.93 | 13.84 | 77.69 | 46.91 | 64.58 | 23.14 | 6.35 | 33.41 | 9.42 | 38.12 | 39.06 | |
| DP14 | 9.52 | 1.98 | 78.99 | 10.27 | 10.74 | 78.10 | 46.59 | 68.13 | 25.06 | 7.01 | 32.54 | 8.98 | 37.04 | 37.93 | |
| DP15 | 9.24 | 1.85 | 79.46 | 10.10 | 10.45 | 78.00 | 49.61 | 65.97 | 26.06 | 6.64 | 33.33 | 8.52 | 37.6 | 38.45 | |
| DP16 | 10.76 | 1.76 | 82.99 | 8.24 | 8.77 | 77.77 | 48.93 | 60.55 | 24.64 | 5.03 | 24.63 | 4.96 | 27.11 | 27.61 | |
| FP (No. p yr−1) | PCS (cm2) | RL (m) | LL (cm) | LW (cm) | LN (No. p yr−1) | HT (m) | LA (cm2) | LAI | DIA (cm) | FI | LAR | FFB (kg palm−1 yr−1) | BN (bunches palm−1 yr−1) | ABW (kg bunch−1) | |
| DP1 | 26.11 | 33.66 | 5.93 | 93.5 | 6.06 | 170.21 | 2.83 | 10.95 | 6.48 | 0.62 | 3.06 | 17.6 | 128.49 | 8.14 | 16.66 |
| DP2 | 24.7 | 32.09 | 5.88 | 105.07 | 5.13 | 172.2 | 2.31 | 10.61 | 6.28 | 0.69 | 3.13 | 17.81 | 136.18 | 9.24 | 14.83 |
| DP3 | 26.35 | 31.88 | 5.39 | 95.7 | 5.4 | 163.7 | 2.1 | 9.67 | 5.72 | 0.67 | 2.85 | 16.51 | 109.63 | 8.43 | 13.38 |
| DP4 | 26.4 | 22.72 | 5.04 | 93.72 | 5.05 | 166.75 | 1.96 | 9.12 | 5.4 | 0.63 | 3.72 | 21.34 | 84.77 | 6.68 | 12.29 |
| DP5 | 27.7 | 24.48 | 5.35 | 91.84 | 4.97 | 163.65 | 2.31 | 8.55 | 5.06 | 0.62 | 3.19 | 18.36 | 107.81 | 8.48 | 13.15 |
| DP6 | 27.65 | 32.81 | 5.73 | 96.47 | 5.52 | 172.5 | 2.42 | 10.48 | 6.2 | 0.68 | 2.98 | 17.19 | 118.8 | 7.6 | 15.95 |
| DP7 | 27.58 | 23.2 | 5.22 | 97.18 | 5.36 | 162.21 | 2.28 | 9.69 | 5.73 | 0.63 | 3.9 | 22.02 | 130.32 | 9.92 | 13.31 |
| DP8 | 26.25 | 31.3 | 5.51 | 92.83 | 5.57 | 170.9 | 2.49 | 10.08 | 5.97 | 0.64 | 3.03 | 17.35 | 126.45 | 8.43 | 15.33 |
| DP9 | 27.2 | 28.18 | 5.75 | 100.55 | 5.39 | 166.75 | 2.37 | 10.31 | 6.1 | 0.6 | 3.5 | 20.37 | 131.78 | 9.28 | 14.94 |
| DP10 | 27.9 | 29.93 | 5.3 | 92.27 | 5.67 | 170.4 | 2.64 | 10.14 | 6 | 0.58 | 3.14 | 18.48 | 118.82 | 7.88 | 15.3 |
| DP11 | 26.5 | 28.73 | 5.59 | 92.82 | 5.6 | 168.6 | 2.7 | 9.99 | 5.91 | 0.59 | 3.25 | 18.7 | 146.15 | 9.5 | 15.73 |
| DP12 | 25.74 | 28.92 | 5.94 | 98.46 | 5.63 | 165.53 | 2.49 | 10.49 | 6.21 | 0.65 | 3.4 | 19.15 | 142.24 | 9.3 | 15.54 |
| DP13 | 28.47 | 26.03 | 5.72 | 99.11 | 5.57 | 168.53 | 2.31 | 10.59 | 6.27 | 0.71 | 3.86 | 21.33 | 144.39 | 10.22 | 14.3 |
| DP14 | 28 | 29.92 | 6.14 | 103.46 | 5.76 | 175.53 | 2.3 | 12.04 | 7.13 | 0.7 | 3.84 | 21.68 | 138.55 | 9.01 | 15.3 |
| DP15 | 26.75 | 30.61 | 6.08 | 98.34 | 5.92 | 174.45 | 2.47 | 11.56 | 6.84 | 0.68 | 3.56 | 20 | 124.38 | 7.92 | 15.7 |
| DP16 | 24.44 | 26.43 | 4.93 | 95.37 | 4.82 | 170.19 | 1.53 | 8.92 | 5.28 | 0.7 | 3.1 | 18.01 | 96.23 | 6.57 | 15.36 |


