1 Introduction
In many groups, including tailed amphibians, decoupling of molecular and morphological evolution (i.e. high level of genetic differentiation that is not paralleled by substantial morphological differentiation with recognisable geographic patterns), has been observed both within and between species [1–3]. Although it is commonly suspected that molecular evolution, including mtDNA, proceeds independently of morphological change, it is important that any association between the morphological and mtDNA evolution rates should be thoroughly tested [4].
It is widely recognized that the European newts (Triturus spp., Lissotriton spp. and Ichthyosaura sp.) offers a unique study system to assess the processes and patterns of many issues of evolutionary biology [5]. As far as the relation between morphology and genetics is concerned, an obvious drawback is that studies of differentiation patterns on these two levels, rigorously tested for the significance of differences, are still missing, especially comparative studies on different morphological datasets. Recently, a study on morphologically and ecologically diversified species of large-bodied crested newts (Triturus cristatus superspecies) revealed congruence between genetic variability and differences in skull shape [6]. Also, other crested newts’ morphological traits such as body size and shape and the number of trunk vertebrae, have been interpreted as reflecting phylogeny [7–9]. In contrast, the phylogenetic substructure of the small-bodied smooth newt (Lissotriton vulgaris) did not coincide with the considerable intraspecific and geographically based morphological disparities [10]. However, there is as-yet scarce relevant data on the medium sized alpine newts [11].
The alpine newt (Ichthyosaura alpestris, Laurenti, 1768), traditionally designated as Triturus alpestris, forms a well-differentiated and strongly supported monophyletic group within the European newt phylogeny [12–14]. Albeit vaguely, the alpine newt is regarded as the most basal species among European newts on the basis of some morphological and mating behaviour characteristics, as well as on molecular sequence data [12,13,15]. The alpine newt is a widespread and ecologically highly flexible newt, especially in terms of breeding site characteristics, which have not undergone ecological specialisation [16]. The alpine newt is considered a polytypic species [16], but the status and nomenclature of its subspecies has a confused taxonomic history due to the lack of well-supported, geographically-based, intraspecific variation patterns in morphology [17]. As in other European newts, morphological and genetical diversities have been described through alpine newt's range, mainly concentrated in the southern part of its distribution, a region where several glacial refugia could have existed.
The Balkan Peninsula has been considered the origin of the alpine newt and the centre of its evolutionary diversification, which includes a great deal of mtDNA-based phylogeographic substructuring [18]. The most distinct lineage, geographically restricted to the Vlasina Region (southeastern Serbia), appears to be an unexpectedly well-diverged basal lineage (Fig. 1) [18]. The most widespread phylogeographic lineage, subendemic to the Balkan Peninsula (hereafter referred to as the Balkan lineage), has two well-differentiated sublineages: the Balkan and the Montenegrin (Fig. 1). Another fairly distinct lineage covers Greece and the adjacent parts of Albania and the FYR of Macedonia (hereafter referred to as the Greek lineage), with additional prominent substructuring including a group of northern populations (hereafter referred to as the Northern Greek sublineage) and southern populations (hereafter referred to as the Southern Greek sublineage) [19]. Such a deep phylogeographic substructure is in contrast with the lack of any geographic pattern in external morphological diversity of the alpine newt on the Balkans [17]. However, a recent investigation of skull shape disparities, based on geometric morphometric analyses within and among the same Balkan phylogenetic lineages, revealed that the dorsal cranium carries a significant phylogenetic signal (i.e., similarity due to shared phylogenetic history), while skull size and ventral skull shape were not phylogenetically informative [11]. Therefore, there is a need for further morphological investigation of the alpine newt in the Balkans aimed at conclusively ascertaining the rate and association of morphological evolution of these lineages.
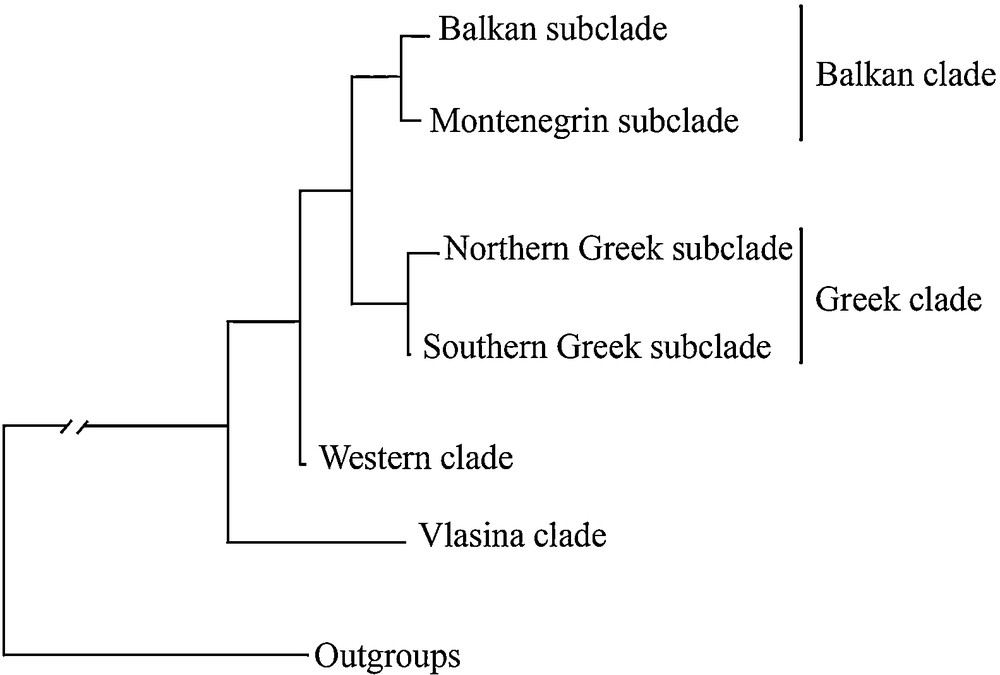
Phylogenetic relationships among the alpine newt lineages based on mtDNA sequences (adapted from Sotiropoulos et al., [18]).
In order to test our main hypothesis that deep phylogeographic pattern is accompanied by extensive morphological differentiation we: (1) test for possible phylogenetic signals in morphological traits; (2) analyse the patterns of variation of morphological traits within and among all previously defined phylogeographic lineages on the Balkans; and (3) estimate the relative evolutionary rates of genetic traits and various morphological traits.
2 Material and methods
2.1 Samples
Our data was obtained from previously analysed populations [11,17,20]. The locations of the 32 studied population samples are presented in Fig. 2 (see Appendix A for the list of population samples, lineage and sublineage affiliations, geographic positions and sample sizes). For lineage and sublineage distribution pattern see Sotiropoulos et al. [18]. Only fully metamorphosed, adult individuals were used. These individuals came from the Herpetological Collections of the Institute for Biological Research “Siniša Stanković” (Belgrade, Serbia) and the Zoological Museum of the University of Athens (Athens, Greece). All specimens were preserved in 70% ethanol for more than five years before morphological analyses were performed.
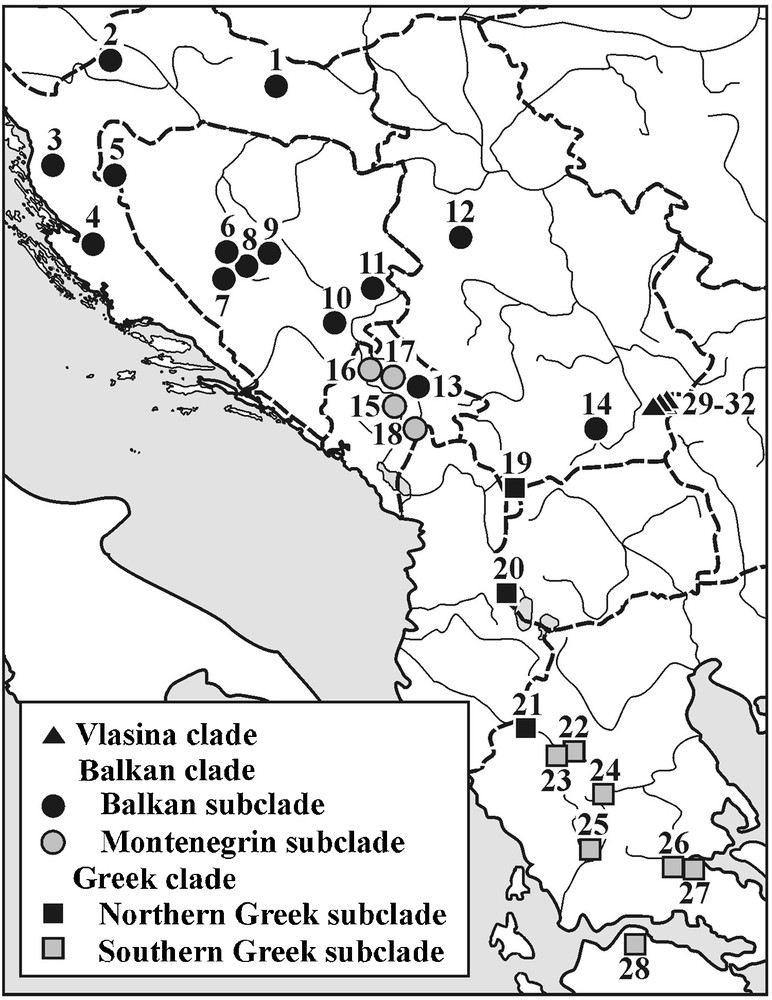
Geographical positions of the analysed population samples and their phylogeographic affiliations.
2.2 Morphological data sets
Morphometric data from 30 populations (1453 specimens) were analysed (see Appendix A for population sample data). Due to significant sexual dimorphism in body size and shape in alpine newts [21], the sexes were analysed separately. Seven body measures were scored: snout-vent length (SVL), interlimb distance (D), head width (HW), head length (HL), forelimb length (FLL), hindlimb length (HLL) and tail length (TL). As size can confound comparisons of variance patterns (i.e., traits with greater means typically have greater variances [22]), the morphometric data were partitioned based on the size and shape sets. Individual size was taken as the geometric mean of all seven body measures for each specimen. Mosimann's ratio method [23] was used to obtain shape variables. Each variable was divided by the geometric mean of all variables for that population. This adjustment removed isometric size but not size-related (allometric) shape.
The qualitative skull data came from 187 specimens from nine population samples that represented all analysed phylogeographic lineages and sublineages (see Appendix A for population sample data). The skulls were cleared with trypsin and KOH and stained with Alizarin red S for bone deposits [24]. Images of ventral and dorsal skulls, with the palate and skull roof positioned parallel to the photographic plane, were obtained with a Sony DSC-F828 digital camera (2592 × 1944 pixels resolution). Skull photographs were used for qualitative trait scoring. In this study, the qualitative skull traits were scored similar to the methods of Bolkay [12] and Herre [25]. For skull analyses, the sexes were pooled because of a lack of significant sexual dimorphism in analysed skull traits (results of the preliminary G test with Williams’ correction). The homologous reference points that are essential for understanding the qualitative descriptions of traits are given in Fig. 3. Thirteen skull traits were scored (see Appendix B and Fig. 3).
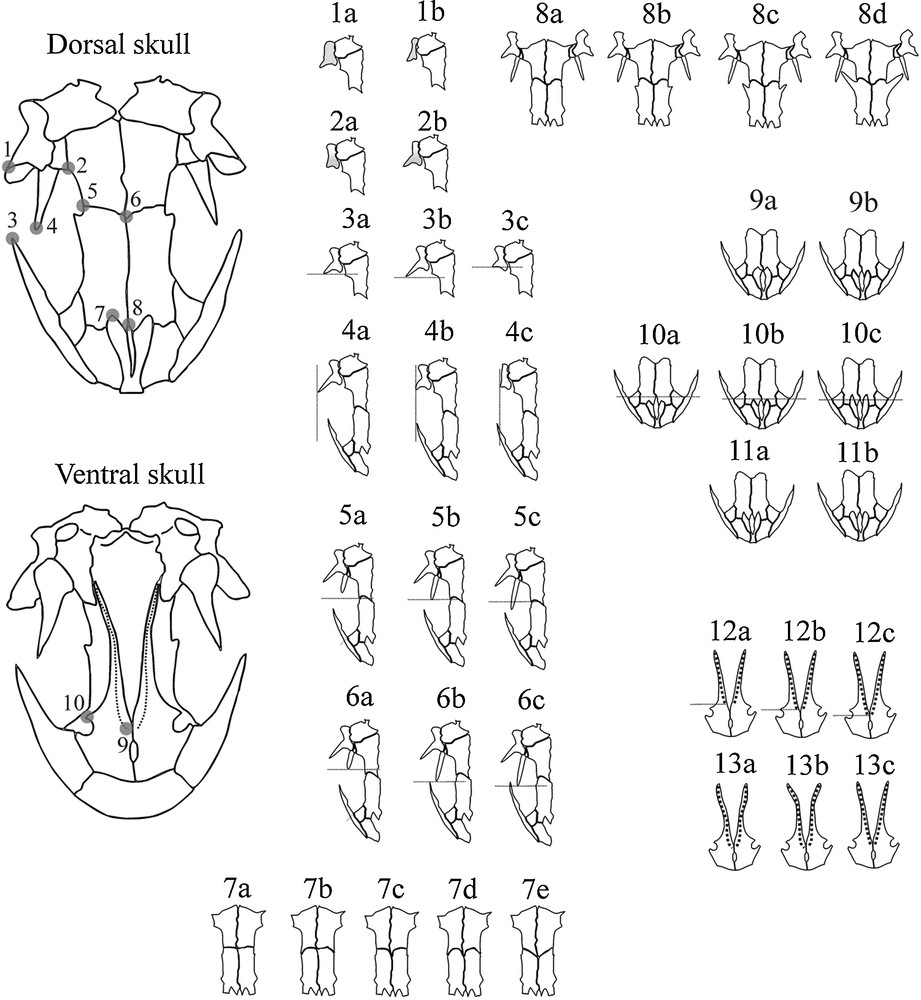
The reference homologous landmark points and the scored qualitative traits. 1. anterior tip of the squamosal; 2. point where the parietal intersects the otic capsule; 3. posterior tip of the maxilla; 4. anterior tip of the pterygoid; 5. lateral point where the frontal intersects the parietal; 6. midpoint where the frontal intersect the parietals; 7. posterior tip of the premaxilla; 8. midpoint where the frontals meet; 9. anterior tip of the vomeral teeth; 10. posterior tip of the choana. For descriptions of the trait states, see Appendix B.
We counted the trunk vertebrae which number has been used in systematics of caudate amphibians quite often [26], especially in crested newts [7]. The number of trunk vertebrae (exclusive of the atlas and sacral vertebra) was counted for 118 specimens belonging to 10 population samples representing all lineages and sublineages (see Appendix A for population sample data). The vertebrae were counted using X-rayed specimens or cleared and stained specimens. The skeletons were cleared with trypsin and KOH and stained with Alizarin red S to visualise bone deposits [24]. The preliminary Mann-Whitney test showed a lack of significant sexual dimorphism in the number of trunk vertebrae, and the sexes were therefore pooled.
For all data sets, we included the appropriate character/lineage ratio so that it adequately reflected the extent of morphological variation in the study system [27].
2.3 Tests for phylogenetic signal in morphological data sets
Since the inter- and intraspecific morphometric data sets may show significant phylogenetic signals [28], we searched for possible signals in the body size, body shape, skull traits, and vertebrae number. As a phylogenetic framework in which to place morphological traits, we used a mitochondrial 16S rRNA/Cyt b phylogeny [18] (see Fig. 1).
To test for phylogenetic signal in size and shape, the canonical procedure that allowed decomposition of quantitative trait variance along the phylogenetic tree [29] was used. Four tests computed from the variance decomposition were used to test the null hypothesis, which was the absence of any phylogenetic dependence: (1) R2Max statistics were used to test the alternative hypothesis that the unique vector of the orthonormal basis explained the significant part of trait variance (punctual effect); (2) S2R2k statistics were used to test the alternative hypothesis that the vectors near the tips or the root (high or low value of S2R2k, respectively) explained the significant part of trait variance; (3) Dmax statistics were used to test the alternative hypothesis that some successive vectors explained a significant part of trait variance; and (4) SCE, which measures the average local variation of the orthogram values, was used to test the alternative hypothesis that there are significant differences in variance explained by vectors and their neighbours (precedent or subsequent). The distribution of the statistics under the null hypothesis and the confidence limits of the cumulative orthograms were built using 999 random permutations of the trait values. The analyses were conducted using the adephylo package [30] in R 2.10.1 [31].
To test for phylogenetic signal in skull traits, and the number of vertebrae, the maximum parsimony ancestral state reconstruction was used. The presence of phylogenetic dependence was tested by reshuffling of terminal taxa 1000 times and confirmed if fewer character changes on a tree have occurred than expected by chance [32]. The majority or “modal” coding was used for coding polymorphism. The analyses were conducted using the Mesquite package [33].
To test for phylogenetic signal in the morphological data, we used the population samples that corresponded to the samples used in a previous study of mtDNA sequence variation [18], which is crucial for the association between mtDNA and morphological variation to be tested rigorously (see Appendix A).
2.4 Tests for variation in morphological data sets
After checking for potential phylogenetic dependence, variation in various morphological traits was studied. In order to reduce the dimensionality for multivariate data sets, we used Principal Component Analysis (PCA) for shape traits set and Detrended Correspondence Analysis (DCA) for skull traits. Data for testing for variations were: (1) sets that contain variables with normal distribution (size, PC scores for shape, DC scores for skull) and (2) discrete variable (number of trunk vertebrae). The main tool for the description of variation pattern of variables with normal distribution was model II nested analysis of variance (PROC NESTED; SAS 9.1) with aim to partition the variance between lineages, among sublineages nested within species and among populations nested within sublineages. Since the sample sizes were not equal, Satterthwaite's approximation was used to test for significance. For the discrete variable, a Mann-Whitney test was used. All analyses were performed using the SAS 9.1 for Windows (SAS Institute Inc., Cary, North Carolina, USA), Statistica 6.0 (StatSoft, Tulsa, Oklahoma, USA), and PC-ORD v 4.25 software [34].
2.5 mtDNA and morphological evolutionary rates
In order to calculate the mtDNA and morphological evolutionary rates, divergence times from the 16S and Cyt b gene phylogenies were used [18]. Evolutionary rates were calculated as distances (standardized values) based on molecules or morphology divided by the corresponding divergence time between groups. Distance standardization were performed in order to be able to compare evolutionary rates, considering that different types of distances between populations were calculated for different data sets (genetic, body size, body shape and skull). The standardized values of the distances were obtained by division of each value by the maximum for the particular distance matrix. Due to this transformation, all distances ranged from zero to one. To avoid division by zero in the evolutionary rate calculations, divergence times with a value of zero were replaced with a value of one year. Due to recent divergence times within sublineages with relatively large estimated error and because of polytomy within some sublineages [18], evolutionary rates were not calculated for populations within the same sublineage. The relations between evolutionary rates of different data sets were evaluated by ANOVA test and Pearson's correlation coefficient.
3 Results
3.1 Phylogenetic signals in morphological traits
For body size and shape, all four statistics tested (R2Max, SkR2k, Dmax, SCE) showed complete absence of any phylogenetic signal (P > 0.05), for both sexes. Similarly, for the skull traits and the number of vertebrae, ancestral state reconstruction and testing for phylogenetic dependence confirmed the absence of phylogenetic signal also.
Since the testing for phylogenetic dependence demonstrated an absence of phylogenetic signal in the analysed morphological traits, there was no need for phylogenetic correction of the morphological data.
3.2 Morphological traits variation patterns
Prior to the nested analyses of variance, all multivariate data sets (body shape and skull traits) were subjected to PCA and DCA to reduce the dimensionality and pinpoint the traits with the most variation.
The seven shape traits were analysed using PCA. The first PC axis explained 42.9% of variation in males and 40.6% in females, while the second PC axis explained 18.1% and 22.5% of variation in males and females respectively. In both sexes, the variables with the highest scores were SVL and D on PC1, while FLL and HLL had the highest scores on PC2 (results not shown).
The states of 13 skull traits were analysed using DCA. The first axis of the DCA for qualitative traits of the skull explained 54.1%, and the second axis explained only 2.1% of the variation. The variables with the highest scores on DC1 were the shape of vomeral teeth and the position of the anterior tip of the squamosal with respect to the point where the parietal intersects the otic capsule. The variable with the highest score on DC2 was the position of the anterior tip of the pterygoid with respect to the posterior tip of the maxilla (results not shown).
In order to analyse the patterns of variation of morphological traits within and among all previously defined phylogeographic lineages on the Balkans, a nested analysis of variance was performed for size, PC scores for shape traits and DC scores for skull traits. At the lineage level, a small amount of no significant variation was found, indicating the absence of a lineage effect on the variation pattern in body size and body shape (Table 1). For skull traits, moderate but insignificant variances at the lineage level were found (Table 2). For body size, body shape and skull traits, the great deal of variation existed within populations (see error values in Tables 1 and 2).
Variance components from a nested analysis of variance in size and in shape (PC1 and PC2 scores). Each component is expressed as the percentage of the total variance (%). PC1 and PC2 are multivariate variables from a Principal Component Analysis. Error variation represents variation within populations plus basic error variance.
| Trait | Between lineages | Among sublineages within lineages | Among populations within sublineages | Error | ||||||||||
| m | f | m | f | m | f | m | f | |||||||
| % | P | % | P | % | P | % | P | % | P | % | P | % | % | |
| Size | 0.0 | ns | 0.0 | ns | 23.0 | 0.0352 | 11.3 | ns | 33.9 | < 0.0001 | 47.8 | < 0.0001 | 41.3 | 40.8 |
| Shape (PC1) | 3.7 | ns | 5.0 | ns | 0.0 | ns | 0.0 | ns | 26.8 | < 0.0001 | 20.5 | < 0.0001 | 69.5 | 74.4 |
| Shape (PC2) | 14.6 | ns | 8.3 | ns | 0.2 | ns | 0.0 | ns | 25.1 | < 0.0001 | 26.5 | < 0.0001 | 60.2 | 65.1 |
Variance components from a nested analysis of variance in skull traits (DC1 and DC2 scores). Each component is expressed as a percentage of the total variance (%). DC1 and DC2 are multivariate variables from a Detrended Correspondence Analysis. Error variation represents variation within populations plus basic error variance.
| Trait | Between lineages | Among sublineages within lineages | Among populations within sublineages | Error | |||
| % | P | % | P | % | P | % | |
| DC 1 | 28.3 | ns | 9.2 | ns | 0.0 | ns | 62.5 |
| DC 2 | 5.9 | ns | 0.0 | ns | 12.3 | 0.0066 | 81.8 |
This study is the first comprehensive study of the variation pattern of number of rib-bearing vertebrae in the alpine newt. All specimens from the Vlasina lineage and the Greek sublineage had 13 trunk vertebrae. The number of trunk vertebrae found in the Balkan lineage was variable: 83.3% individuals had 12 trunk vertebrae, 8.3% (four individuals) had 11 vertebrae and the remaining four individuals (8.3%) had 13 trunk vertebrae (see Appendix A). Specimens from the Vlasina and Greek lineages showed a significant difference in the number of trunk vertebrae compared to the Balkan lineage (Mann Whitney, P < 0.001). The Balkan sublineages could not be distinguished by the number of trunk vertebrae (Mann Whitney, P > 0.05); 75% individuals of the Balkan sublineage and 89.3% individuals of the Montenegrin sublineage had 12 trunk vertebrae.
3.3 mtDNA and morphological evolutionary rates
An explicit pattern of variation of the evolutionary rates could not be found because of the extensive variability in all data sets (Table 3). In general, the relative rates of change for all morphological traits were similar to the rate of mtDNA change (ANOVA test, P > 0.05). Four morphological data sets (size in males, size in females, shape in males, shape in females) had larger number of evolutionary rates below the corresponding mtDNA rate. Only traits of skull had larger number of evolutionary rates above the corresponding mtDNA rate (Table 3). There was no correlation between the mtDNA and morphological rates, while among all morphological data sets correlation of rates were significant (data not shown).
Genetic and morphological evolutionary rates of the alpine newt's population pairs.
| Genetic | Size (m) | Size (f) | Shape (m) | Shape (f) | Skull | |
| Velouchi Mt./Kutarevo | 0.21 | 0.08 | 0.06 | 0.21 | 0.37 | 0.28 |
| Katavothra/Kutarevo | 0.21 | 0.02 | 0.07 | 0.15 | 0.24 | 0.28 |
| Manito L./Kutarevo | 0.20 | 0.46 | 0.57 | 0.26 | 0.26 | 0.54 |
| Bukumirsko L./Kutarevo | 0.21 | 0.27 | 0.24 | 0.28 | 0.23 | 0.54 |
| Vlasina Stojk./Kutarevo | 0.18 | 0.03 | 0.02 | 0.04 | 0.07 | 0.13 |
| Šara/Kutarevo | 0.20 | 0.13 | 0.27 | 0.13 | 0.05 | 0.28 |
| Mlačiške M./Kutarevo | 0.18 | 0.09 | 0.04 | 0.13 | 0.09 | 0.16 |
| Velouchi Mt./Prokoško L. | 0.20 | 0.37 | 0.35 | 0.10 | 0.34 | 0.32 |
| Katavothra/Prokoško L. | 0.20 | 0.28 | 0.37 | 0.10 | 0.17 | 0.32 |
| Manito L./Prokoško L. | 0.15 | 0.12 | 0.01 | 0.72 | 0.65 | 0.63 |
| Bukumirsko L./Prokoško L. | 0.16 | 0.31 | 0.34 | 0.64 | 0.51 | 0.63 |
| Vlasina Stojk./Prokoško L. | 0.17 | 0.11 | 0.16 | 0.13 | 0.09 | 0.16 |
| Šara/Prokoško L. | 0.18 | 0.16 | 0.03 | 0.05 | 0.12 | 0.28 |
| Mlačiške M./Prokoško L. | 0.17 | 0.05 | 0.10 | 0.12 | 0.15 | 0.18 |
| Manito L./Velouchi Mt. | 0.17 | 0.31 | 0.35 | 0.25 | 0.28 | 0.28 |
| Bukumirsko L./Velouchi Mt. | 0.17 | 0.21 | 0.18 | 0.25 | 0.29 | 0.23 |
| Vlasina Stojk./Velouchi Mt. | 0.17 | 0.07 | 0.01 | 0.10 | 0.12 | 0.11 |
| Šara/Velouchi Mt. | 0.15 | 0.48 | 0.76 | 0.24 | 0.71 | 0.54 |
| Mlačiške M./Velouchi Mt. | 0.17 | 0.13 | 0.07 | 0.07 | 0.07 | 0.13 |
| Manito L./Katavothra | 0.17 | 0.22 | 0.37 | 0.14 | 0.15 | 0.23 |
| Bukumirsko L./Katavothra | 0.17 | 0.12 | 0.19 | 0.13 | 0.10 | 0.28 |
| Vlasina Stojk./Katavothra | 0.17 | 0.02 | 0.01 | 0.06 | 0.04 | 0.11 |
| Šara/Katavothra | 0.15 | 0.27 | 0.80 | 0.09 | 0.49 | 0.54 |
| Mlačiške M./Katavothra | 0.17 | 0.08 | 0.07 | 0.03 | 0.03 | 0.13 |
| Vlasina Stojk./Manito L. | 0.17 | 0.08 | 0.16 | 0.02 | 0.04 | 0.11 |
| Šara/Manito L. | 0.16 | 0.10 | 0.02 | 0.18 | 0.16 | 0.28 |
| Mlačiške M./Manito L. | 0.17 | 0.02 | 0.10 | 0.07 | 0.05 | 0.16 |
| Vlasina Stojk./Bukumirsko L. | 0.16 | 0.04 | 0.08 | 0.03 | 0.02 | 0.11 |
| Šara/Bukumirsko L. | 0.16 | 0.00 | 0.15 | 0.14 | 0.16 | 0.28 |
| Mlačiške M./Bukumirsko L. | 0.16 | 0.03 | 0.02 | 0.07 | 0.05 | 0.16 |
| Šara/Vlasina | 0.15 | 0.03 | 0.15 | 0.06 | 0.05 | 0.11 |
| Mlačiške M./Šara | 0.15 | 0.03 | 0.09 | 0.07 | 0.12 | 0.16 |
| Median | 0.17 | 0.11 | 0.13 | 0.13 | 0.14 | 0.26 |
| Min | 0.15 | 0.00 | 0.01 | 0.02 | 0.02 | 0.11 |
| Max | 0.21 | 0.48 | 0.80 | 0.72 | 0.71 | 0.63 |
| + | 10 | 12 | 8 | 11 | 20 | |
| - | 22 | 20 | 24 | 21 | 12 |
4 Discussion
Our data suggest that mtDNA evolution is strongly disassociated from the morphological evolution in the alpine newts from the Balkan Peninsula. The patterns of morphological differentiation are highly redundant across phylogenetic lineages with variation clearly apportioned within rather than between all recognised phylogenetic lineages. Similar patterns have been previously found for the ventral skull portions [11].
Dissociation of mtDNA and morphological evolution could be explained by gene introgression and/or natural selection [10,35]. In such cases, if there is directional selection, a high rate of morphological disparity is expected. Directional selection may also blur the phylogenetic signal, resulting in convergent morphotypes. On the other hand, if there is stabilising selection, morphologically cryptic lineages are expected to appear, an occurrence that appeared to be confirmed in the case of the alpine newt.
Given the lack of geographical structuring in morphology, the observed patterns of morphological variation could be explained by spatial, local environmental factors. Gene flow between adjacent populations of the alpine newt is sufficient low, due to high philopatry and poor dispersal ability [36] to maintain phenotypic differences under selection. In such cases, adaptations are more easily fixed when genetic structure is strong [37] which is the case in the alpine newt [18,19,38]. In other words, our results indicate an in situ morphological diversification of the alpine newt across the Balkans, i.e. morphological differentiation in the alpine newts appears at the smallest geographical scale, at the level of local populations. This result is particularly noteworthy in the context of other two European newts groups which are generally known for their large morphological diversity with more diverse intergroup morphological traits which have been the subject of speciation processes. This diversity is taxonomically appreciated at the species and subspecies levels. Thus, regarding morphology, in the big-bodied crested newts (Triturus cristatus superspecies) species differ mostly in the ecologically related traits, while in the small bodied smooth newt (Lissotriton spp.) subspecies differ mostly in elaborated secondary male sexual integumental traits.
Some osteological characteristics (qualitative skull traits and the number of trunk vertebrae) have some discriminative value in terms of among-lineage comparisons. As envisioned previously by geometric morphometrics, individuals of the Greek and Vlasina lineages proved to be the most divergent in cranial morphology with respect to other phylogeographic entities [11].
The branch leading to the Vlasina lineage is relatively long before diversifying into other alpine newt taxa [18], suggesting that the Vlasina populations might have been restricted to a small mountainous area, lacking any success in long-term colonisation beyond this habitat. In regard to alpine newt's evolutionary history, these suggest that Pleistocene glaciations might have allowed the preservation at least part of the ancestral genetic diversity in the alpine newt which can be designated as “sanctuary species” [39]. It might be a case of morphological diversity as well. If so, the alpine newt from the Balkan Peninsula offers a striking case of possible phenotypic stasis in restricted area over a very long period of evolutionary time. As much of the alpine newt's lineage-level disparity in the external morphology was almost cryptic, most likely due to the long-term stability of the ecological niche, it is possible that cladogenesis occurred through vicariant geographic processes followed by range shifts without obvious adaptive divergences [40]. The long-term stability of an ecological niche has profound consequences with respect to increasing lineage diversity because it causes lineages to track their preferred habitats in the face of environmental change, promoting vicariant fragmentation and the formation of new evolutionary lineages.
Regarding evolutionary rates, the lack of relationship between genetics and morphology has sometimes been interpreted in terms of differences in molecular and morphological evolutionary rates [41]. In our study, such generalisation has not been proved, as the relative rates of change for morphological traits are on average similar to the rate of mtDNA changes. Interestingly enough, to date few studies have revealed such correlation between genetic and morphological rates of change, especially in animals [42].
In conclusion, our results imply that the deep genetic diversification is not accompanied by extensive phenotypic divergence in the alpine newt as far as Balkan alpine newts are concerned, as the mtDNA and morphological analyses provide two qualitatively different data sets although the rates of mtDNA and morphological changes were similar. It seems that alpine newt evolution on the Balkans took place largely through differentiation at the mtDNA level rather than through morphological or, as it seems, ecological diversification as well. As noted above, in contrast to other European newts, which are characterised by adaptive radiations (i.e. high rate of morphological change is following by taxa diversification), in the alpine newt there is a lack of geographical structuring in morphology. To date, available evidence suggests a non-adaptive morphological evolution for the alpine newt or its morphological cryptosis due to similar developmental and evolutionary constraints on body form.
To better understand extrinsic and intrinsic constraints on phenotypic evolution, the study of extant species that exhibit both deep genetic divergence and negligible morphological differentiation is promising. Apparently, the alpine newt offers such a study system.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
Helpful input has been provided by Ana Ivanović and the two reviewers. This study was partially funded with financial support from the Serbian Ministry of Education and Science (project No. 173043).
Appendix A
Location data (name and position) and the number of specimens analysed in various morphological studies (the number of trunk vertebrae is given in parentheses) and for a DNA study. m–males; f–females; *–larvae and juveniles; B&H–Bosnia and Herzegovina.
| Morphometry | Cranium (q. traits) | Trunk vertebrae | mtDNA analyses | Country | Coordinates | Altitude | ||||||
| m | f | m | f | m | f | |||||||
| Balkan lineage | ||||||||||||
| Balkan sublineages | ||||||||||||
| 1 | Jankovac, Mt. Papuk | 18 | 19 | + | Croatia | 45° 31’ N | 17° 41’ E | 457 | ||||
| 2 | Peščenica, Velika Gorica | 16 | 19 | Croatia | 45° 36’ N | 16° 10’ E | 105 | |||||
| 3 | Kutarevo, Lika | 31 | 41 | 12 | 12 | + | Croatia | 44° 51’ N | 15° 10’ E | 560 | ||
| 4 | Laudanovi Gaj, Krbavsko Polje | 26 | 28 | Croatia | 44° 38’ N | 15° 38’ E | 630 | |||||
| 5 | Ušljebrka, Žegar | 29 | 34 | + | Croatia | 44° 09’ N | 15° 51’ E | 80 | ||||
| 6 | Rastičevsko Lake, Blagaj | 43 | 23 | B&H | 44° 02’ N | 17° 13’ E | 1180 | |||||
| 7 | Karaizovići, Glamočko Polje | 36 | 41 | B&H | 44° 02’ N | 16° 50’ E | 890 | |||||
| 8 | Šupljica, Kupreska Vrata | 26 | 46 | B&H | 43° 59’ N | 17° 16’ E | 1350 | |||||
| 9 | Prokoško Lake, Mt. Vranica | 45 | 111 | 9 | 16 | 1 (13) 7 (12) 2 (11) | 2 (13) 8 (12) | + | B&H | 43° 57’ N | 17° 45’ E | 1635 |
| 10 | Gornje Bare, Mt. Zelengora | 37 | 25 | B&H | 43° 22’ N | 18° 30’ E | 1650 | |||||
| 11 | Rogatica, Seljani | 18 | 19 | + | B&H | 43° 48’ N | 19° 05’ E | 820 | ||||
| 12 | Valjevo, Joševa | 18 | 17 | + | Serbia | 44° 22’ N | 19° 50’ E | 345 | ||||
| 13 | Ursulovačko Lake, Mt. Bjelasica | 11 | 25 | Montenegro | 42° 52’ N | 19° 44’ E | 1760 | |||||
| 14 | Vranje, Mt. Sveti Ilija | 43 | 33 | + | Serbia | 42° 36’ N | 21° 51’ E | 1120 | ||||
| Total | 397 | 481 | 21 | 28 | 10 | 10 | ||||||
| Montenegrin sublineage | ||||||||||||
| 15 | Zminje Lake, Mt. Durmitor | 45 | 54 | Montenegro | 43° 09’N | 19° 04’ E | 1495 | |||||
| 16 | Manito Lake, Lukavica | 49 | 46 | 15 | 16 | 10 (12) | 8 (12) 1 (13) | Montenegro | 42° 48’ N | 19° 14’ E | 1773 | |
| 17 | Zminičko Lake, Mt. Sinjavina | 19 | 10 | + | Montenegro | 42° 59’ N | 19° 15’ E | 1285 | ||||
| 18 | Bukumirsko Lake | 18 | 14 | 11 | 10 | 2 (11)*; 7 (12)* | Montenegro | 42° 36’ N | 19° 33’ E | 1440 | ||
| Total | 131 | 124 | 26 | 26 | 10 | 9 + 9* | ||||||
| Greek lineage | ||||||||||||
| Northern Greek sublineage | ||||||||||||
| 19 | Donje Ravne Mlake, Mt. Šara | 17 | 20 | 10 | 10 | 10 (13) | 10 (13) | + | Serbia | 41° 54’ N | 20° 32’ E | 2100 |
| 20 | Podgorečko Lake, Mt. Jablanica | 18 | 21 | + | FYR Macedonia | 41° 15’ N | 20° 32’ E | 1870 | ||||
| 21 | Smolikas Mt. | 10 | 11 | + | Greece | 40° 05’ N | 20° 54’ E | 2180 | ||||
| Total | 45 | 52 | 10 | 10 | 10 | 10 | ||||||
| Southern Greek sublineage | ||||||||||||
| 22 | Zygos Mt. | 8 | 14 | + | Greece | 39° 53’ N | 21° 17’ E | 1450 | ||||
| 23 | Limni Pigon Aoou | 4 (13) | 4 (13) | + | Greece | 39° 49’ N | 21° 08’ E | 1400 | ||||
| 24 | Kerketio Mt. | 8 | 9 | + | Greece | 39° 31’ N | 21° 31’ E | 1050 | ||||
| 25 | Velouchi Mt. | 8 | 9 | 8 | 9 | 8 (13) | 9 (13) | + | Greece | 38° 57’ N | 21° 49’ E | 1850 |
| 26 | Katavothra, Oeta Mt. | 11 | 8 | 11 | 8 | 11 (13) | 8 (13) | + | Greece | 38° 46’ N | 22° 19’ E | 1550 |
| 27 | Kallidromo Mt. | 14 | 14 | + | Greece | 38° 45’ N | 22° 33’ E | 950 | ||||
| 28 | Panachaiko Mt., Rakita Plateau | 3 (13) | 3 (13) | + | Greece | 38° 10’ N | 21° 54’ E | 1050 | ||||
| Total | 49 | 54 | 19 | 17 | 26 | 24 | ||||||
| Vlasina lineage | ||||||||||||
| 29 | Klisura | 12 | 12 | + | Serbia | 42° 46’ N | 22° 25’ E | 857 | ||||
| 30 | Selište | 14 | 19 | Serbia | 42° 50’ N | 22° 12’ E | 780 | |||||
| 31 | Stojkovića Mahala | 19 | 20 | 10 | 10 | 10 (13) | 10 (13) | + | Serbia | 42° 44’ N | 22° 19’ E | 1340 |
| 32 | Mlačiške Meane | 10 | 14 | 4 | 6 | 10 (13) | 14 (13) | + | Serbia | 42° 46’ N | 22° 14’ E | 1294 |
| Total | 55 | 65 | 14 | 16 | 20 | 24 | ||||||
| Grand Total | 677 | 776 | 90 | 97 | 162 |
Appendix B Qualitative skull traits
1. Squamosal shape - (a) short, robust; (b) long, fragile; 2. Relation of the anterior and posterior parts of the squamosal - (a) anterior and posterior parts have the same width, (b) the anterior is wider than the posterior; 3. Position of the anterior tip of the squamosal with respect to the point where the parietal intersects the otic capsule (1 vs. 2) - (a) on the same level as the point where the parietal intersects the otic capsule; (b) extends past the point where the parietal intersects the otic capsule; (c) does not reach the point where the parietal intersects the otic capsule; 4. Position of the anterior tip of the squamosal with respect to the posterior tip of the maxilla (1 vs. 3) - (a) more laterally (prominent) than the posterior tip of the maxilla; (b) on the same level as the posterior tip of the maxilla; (c) the posterior tip of the maxilla more laterally (prominent) than the anterior tip of the squamosal; 5. Position of the anterior tip of the pterygoid with respect to the lateral point where the frontal intersects the parietal (4 vs. 5) - (a) does not reach the lateral point where the frontal intersects the parietal (pterygoid short); (b) on the same level as the lateral point where the frontal intersects the parietal; (c) goes over the lateral point where the frontal intersects the parietal (pterygoid long); 6. Position of the anterior tip of the pterygoid with respect to the posterior tip of the maxilla (4 vs. 3) - (a) does not reach the posterior tip of the maxilla; (b) on the same level as the posterior tip of the maxilla; (c) extends past the posterior tip of the maxilla; 7. Shape of the posterior margin of the frontals–sutura coronalis (5 vs. 6) - (a) oblate (points 5 and 6 on the same level); (b) concave (point 6 frontal with respect to point 5); (c) concave (points 5 and 6 on the same level); (d) V shape (point 6 frontal with respect to point 5); 8. Size of the processus postfrontalis - (a) processus postfrontalis absent; (b) very short processus postfrontalis; (c) short processus postfrontalis; (d) long processus postfrontalis; 9. Contact of the premaxillary pars dorsalis - (a) posterior contact; (b) no posterior contacting; 10. Position of the posterior tip of the premaxilla with respect to the midpoint where the frontal intersects (7 vs. 8) - (a) does not reach the midpoint where the frontal intersects (short premaxillae); (b) on the same level as the midpoint where the frontal intersects; (c) extends past the midpoint where the frontal intersects (long premaxillae); 11. Shape of the posterior tip of the premaxilla - (a) sharp; (b) rounded; 12. Position of the anterior tip of the vomeral teeth with respect to the posterior tip of the choana (9 vs. 10)–(a) does not reach the posterior tip of the choana; (b) on the same level as the posterior tip of the choana; (c) extends past the posterior tip of the choana; 13. Shape of the vomeral teeth - (a) “wine glass” form; (b) arch form; (c) straight form.


