1 Introduction
Heavy metal P1B-ATPases (HMA proteins) transport heavy metals through biological membranes via an ATP-dependent process. HMA genes constitute a multigene family, which counts eight members in Arabidopsis thaliana [1]. HMA proteins display very strong specificities with respect to the metals that they transport. In A. thaliana, AtHMA2, AtHMA3 and AtHMA4 are involved in zinc/cadmium/lead/cobalt transport whereas AtHMA5, AtHMA6, AtHMA7 and AtHMA8 are involved in copper transport (see [2] for a review). AtHMA1, which occupies an intermediate position between the zinc/cadmium/lead/cobalt- and the copper-transporting HMAs on the phylogenetic tree [1,3,4], can transport copper but also zinc/cadmium/cobalt as well as calcium [5].
The functional roles of the zinc/cadmium/lead/cobalt transporting ATPases are diverse. AtHMA1 is located on the chloroplast envelope and is involved both in copper loading into the chloroplast and in zinc detoxification [6,7]. In A. thaliana, AtHMA2 and AtHMA4 play a critical role in the zinc and cadmium translocation from the roots to the shoot [8–11]. AtHMA2 and AtHMA4 are located on the plasmalemma of stellar cells and regulate the zinc and cadmium contents in vessels. When the hma2 mutation is present in an hma4 mutant background, it amplifies the phenotypic change induced by the single hma4 mutation [9,11], indicating that HMA2 is functionally redundant with HMA4. In Brassicaceae and in particular in plant species showing very high tolerance to zinc and cadmium such as Arabidopsis halleri and Noccaea caerulescens, HMA4 also appears to be a major determinant of zinc and cadmium tolerance [12–15]. In A. halleri for instance, HMA4 is co-localised with a major QTL that controls both zinc and cadmium tolerance along with the accumulation of these two elements [12,15,16]. HMA4 is triplicated in this species, the three paralogues being highly expressed [13]. RNAi-mediated inactivation of HMA4 in A. halleri was shown to induce a decrease in zinc and cadmium tolerance as well as a decrease in zinc and cadmium accumulation in shoots [13]. In N. caerulescens, HMA4 was very recently shown to be quadruplicated [17].
Data are also available in rice where OsHMA3 seems to be the major determinant controlling cadmium – but not zinc – translocation from the roots to the shoot [18,19]. However, OsHMA3 is involved in a completely different mechanism than AtHMA2 or AtHMA4. Indeed, OsHMA3 is not located on the plasmalemma of stellar cells but it is present on the tonoplast of root cells and controls the loading of cadmium into the vacuole. Amazingly, there is also a Zn/Cd/Pb/Co P1B-ATPase (named AtHMA3) that is located on the tonoplast of root cells and involved in the cadmium, lead and cobalt transport into the vacuole in A. thaliana, but in contrast to OsHMA3, it has no demonstrated impact on the root to shoot translocation of cadmium [20].
The OsHMA3, AtHMA2, AtHMA3 and AtHMA4 transporters do not display exactly the same ion selectivity [10,18–21]. For instance, AtHMA3 transports cadmium, lead and cobalt while AtHMA2 and AtHMA4 transport zinc and cadmium. The molecular bases of this difference in selectivity are still unknown. Also, the protein structures of these transporters are different. The Zn/Cd/Pb/Co P1B-ATPases possess a transmembrane domain with eight transmembrane helices in their N-terminal part and a cytosolic domain in their C-terminal part [3]. Whereas the transmembrane domains are very similar between HMA proteins, the C-terminal domains are very different both in length and in amino acid sequence. AtHMA3 displays a particularly short C-terminal domain compared to OsHMA3, AtHMA2 and AtHMA4 [3,18–20], and this may explain the functional differences observed between these transporters. Indeed, the cytosolic domain is supposed to play a regulatory role with respect to the enzyme activity [22] but it could also play many other roles, including the subcellular targeting [4,23].
The functional differences revealed from the comparative analysis of the different Zn/Cd/Pb/Co P1B-ATPases present in A. thaliana, as well as from the comparison between the A. thaliana and rice heavy metal ATPases suggest that it will not be easy to directly transfer the acquired knowledge to other plant species, for instance with the idea to monitor the accumulation of heavy metals in these species and breed varieties well suited for phytoremediation approaches. In particular, given the functional diversity that the HMA2, HMA3 and HMA4 genes actually display in A. thaliana, it is of great importance to determine the appropriate orthologues of the HMA genes in every plant species of interest. While developing such a project, we observed that the phylogenetic relationships linking the Zn/Cd/Pb/Co P1B-ATPases of different plant species were very particular. This report thus presents the analysis of the phylogenetic relationships linking the genes encoding Zn/Cd/Pb/Co P1B-ATPases from different plant species. Our results suggest that the orthology relationships are clear considering species belonging to a same botanical family, but that these relationships cannot be clearly established when considering plants belonging to different botanical families.
2 Materials and methods
2.1 Identification of sequences encoding heavy metal P1B-ATPases
Nucleotide sequences of heavy metal P1B-ATPases showing orthology to already identified heavy metal P1B-ATPases were extracted from the Genbank sequence databases following similarity analyses performed using the TBLASTN program with the filtering removed. The sequence databases that were analysed were the ‘non-redundant’ (nr) database, the ‘high throughput genome sequences’ (htgs) database, the ‘whole-genome shotgun reads’ (wgs) database and the ‘NCBI genome’ (chromosome) database. The analyses were completed considering the sequences available in databases at the end of April 2011. The set of sequences that we obtained is extensively described in Table 1.
HMA1-2-3-4 sequences found in the public databases at the end of April 2011.
| Species | Family | Gene name | Status of the sequence | Protein / C-terminal tail length (in amino acid) | Accession number of the nucleotide sequence | Reannotated sequence |
| A. halleri | Brassicaceae | AhHMA1 | Partial | 182 | AJ580403 | No |
| A. halleri | Brassicaceae | AhHMA3 | Complete | 757 / 58 | AJ556182 | No |
| A. halleri | Brassicaceae | AhHMA4-1 | Complete | 1161 / 458 | EU382073 | No |
| A. halleri | Brassicaceae | AhHMA4-2 | Complete | 1161 / 458 | EU382072 | No |
| A. halleri | Brassicaceae | AhHMA4-3 | Complete | 1161 / 460 | EU382072 | No |
| A. lyrata | Brassicaceae | AlHMA1-1 | Complete | 826 / 38 | XM_002866914 | No |
| A. lyrata | Brassicaceae | AlHMA1-2 | Complete | 808 / 38 | ADBK01000683 | Yes |
| A. lyrata | Brassicaceae | AlHMA2 | Complete | 944 / 250 | XM_002867321 | No |
| A. lyrata | Brassicaceae | AlHMA3 | Complete | 757 / 58 | XM_002867320 | No |
| A. lyrata | Brassicaceae | AlHMA4-1 | Complete | 1275 / 472 | ADBK01000666 | Yes |
| A. lyrata | Brassicaceae | AlHMA4-2 | Partial | 998 / 487 | ADBK01000159 | Yes |
| A. thaliana | Brassicaceae | AtHMA1 | Complete | 819 / 35 | NM_119890 | No |
| A. thaliana | Brassicaceae | AtHMA2 | Complete | 951 / 257 | AY434728 | No |
| A. thaliana | Brassicaceae | AtHMA3 | Complete | 760 / 59 | AY055217 | No |
| A. thaliana | Brassicaceae | AtHMA4 | Complete | 1172 / 469 | AF412407 | No |
| B. distachyon | Poaceae | BdHMA1 | Complete | 819 / 40 | ADDN01000155 | Yes |
| B. distachyon | Poaceae | BdHMA2 | Complete | 1038 / 335 | ADDN01000162 | Yes |
| B. distachyon | Poaceae | BdHMA3 | Complete | 819 / 103 | ADDN01000328 | Yes |
| B. rapa | Brassicaceae | BrHMA1-1 | Complete | 818 / 37 | FP236818 | Yes |
| B. rapa | Brassicaceae | BrHMA1-2 | Complete | 774 / 4 | AC232485 | Yes |
| B. rapa | Brassicaceae | BrHMA2 | Complete | 905 / 208 | AC240993 | Yes |
| B. rapa | Brassicaceae | BrHMA3-1 | Complete | 758 / 59 | FP017269 | Yes |
| B. rapa | Brassicaceae | BrHMA3-2 | Complete | 764 / 64 | AC241039 | Yes |
| B. rapa | Brassicaceae | BrHMA4-1 | Complete | 872 / 176 | AC232536 | Yes |
| B. rapa | Brassicaceae | BrHMA4-2 | – | FP102280 | Pseudogene | |
| C. papaya | Caricaceae | CpHMA1 | Complete | 830 / 40 | ABIM01002425 | Yes |
| C. papaya | Caricaceae | CpHMA-A | Complete | 1014/316 | ABIM01016748 | Yes |
| C. sativus | Cucurbitaceae | CsHMA1 | Complete | 823 / 38 | ACHR01015552 | Yes |
| C. sativus | Cucurbitaceae | CsHMA-A | Complete | 888 / 176 | ACHR01010422 | Yes |
| C. sativus | Cucurbitaceae | CsHMA-B | Complete | 1231 / 528 | ACHR01008892 | Yes |
| E. parvulum | Brassicaceae | EpHMA1-1 | Complete | 822 / 37 | AFAN01000040 | Yes |
| E. parvulum | Brassicaceae | EpHMA1-2 | Complete | 824 / 35 | AFAN01000016 | Yes |
| E. parvulum | Brassicaceae | EpHMA2 | Complete | 812 / 216 | AFAN01000039 | Yes |
| E. parvulum | Brassicaceae | EpHMA3 | Complete | 760 / 61 | AFAN01000039 | Yes |
| E. parvulum | Brassicaceae | EpHMA4 | Complete | 1310 / 606 | AFAN01000014 | Yes |
| F. vesca | Rosaceae | FvHMA1 | Complete | 874 / 43 | AEMH01014081 | Yes |
| F. vesca | Rosaceae | FvHMA-A | Complete | 1070 / 374 | AEMH01012163 | Yes |
| F. vesca | Rosaceae | FvHMA-B | Complete | 835 / 125 | AEMH01014150 | Yes |
| G. max | Fabaceae | GmHMA1-1 | Complete | 817 / 40 | ACUP01009874 | Yes |
| G. max | Fabaceae | GmHMA1-2 | Complete | 823 / 40 | ACUP01003074 | Yes |
| G. max | Fabaceae | GmHMA-A | Complete | 885 / 193 | ACUP01005112 | Yes |
| G. max | Fabaceae | GmHMA-B | Complete | 1096 / 401 | ACUP01007321 | Yes |
| G. max | Fabaceae | GmHMA-C | Complete | 807 / 114 | ACUP01009759 | Yes |
| G. max | Fabaceae | GmHMA-D | – | ACUP01008779 | Pseudogene | |
| H. incana | Brassicaceae | HiHMA4 | Partial | 288 | HQ398195 | No |
| H. vulgare | Poaceae | HvHMA1-1 | Complete | 828 / 40 | AK374806 | No |
| H. vulgare | Poaceae | HvHMA1-2 | Complete | 774 / 39 | AK358556 | No |
| H. vulgare | Poaceae | HvHMA2 | Complete | 1009 / 307 | AK363365 | No |
| H. vulgare | Poaceae | HvHMA3 | Complete | 838 / 121 | AK369525 | No |
| J. curcas | Euphorbiaceae | JcHMA-A | Partial | 835 / 268 | BABX01026037 | Yes |
| L. sativa | Asteraceae | LsHMA-A | Partial | 223 | FN985047 | No |
| L. sativa | Asteraceae | LsHMA-B | Partial | 171 | FN985050 | Yes |
| M. truncatula | Fabaceae | MtHMA-A | Complete | 829 / 138 | AC130275 | Yes |
| M. truncatula | Fabaceae | MtHMA-B | Complete | 1033 / 335 | AC135313 | Yes |
| N. caerulescens | Brassicaceae | NcHMA4 | Complete | 1186 / 479 | AJ567384 | No |
| N. tabacum | Nicotianeae | NtHMA-A | Complete | 1403 / 703 | HB441191 | No |
| N. tabacum | Nicotianeae | NtHMA-B | Complete | 1294 / 594 | HB441235 | No |
| O. glaberrima | Poaceae | OgHMA1 | Complete | 822 / 40 | ADWL01012602 | Yes |
| O. glaberrima | Poaceae | OgHMA2 | Complete | 1068 / 372 | ADWL01012616 | Yes |
| O. glaberrima | Poaceae | OgHMA3 | Complete | 1004 / 274 | ADWL01013846 | Yes |
| O. sativa | Poaceae | OsHMA1 | Complete | 822 / 40 | NM_001064952 | No |
| O. sativa | Poaceae | OsHMA2 | Complete | 1067 / 371 | HQ646362 | No |
| O. sativa | Poaceae | OsHMA3 | Complete | 1004 / 274 | AB557931 | No |
| P. dactylifera | Arecaceae | PdHMA-A | Partial | 618 | ACYX02014453 | Yes |
| P. dactylifera | Arecaceae | PdHMA-B | Partial | 602 / 220 | ACYX02046688 | Yes |
| P. dactylifera | Arecaceae | PdHMA-C | Complete | 776 / 77 | ACYX02004700 | Yes |
| P. glauca | Pinaceae | PgHMA1 | Partial | 210 | BT106845 | No |
| P. glauca | Pinaceae | PgHMA-A | Partial | 324 | BT102415 | No |
| P. glauca | Pinaceae | PgHMA-B | Partial | 282 | BT119672 | No |
| P. persica | Rosaceae | PpHMA1 | Complete | 824 / 57 | AEKW01002839 | Yes |
| P. persica | Rosaceae | PpHMA-A | Complete | 1067 / 366 | AEKW01008176 | Yes |
| P. trichocarpa | Salicaceae | PtHMA1-1 | Complete | 832 / 40 | AC210508 | Yes |
| P. trichocarpa | Salicaceae | PtHMA1-2 | – | AARH01002640 | Pseudogene | |
| P. trichocarpa | Salicaceae | PtHMA4 | Complete | 1145 / 443 | AARH01002993 | Yes |
| P. trichocarpa | Salicaceae | PtHMA-B | – | AARH01008666 | Pseudogene | |
| R. communis | Euphorbiaceae | RcHMA1 | Complete | 820 / 40 | XM_002524881 | No |
| R. communis | Euphorbiaceae | RcHMA-A | Partial | > 933 / 253 | XM_002532190 | Yes |
| S. bicolor | Poaceae | SbHMA1 | Complete | 828 / 40 | ABXC01006890 | Yes |
| S. bicolor | Poaceae | SbHMA2 | Complete | 1069 / 369 | XM_002438908 | No |
| S. bicolor | Poaceae | SbHMA3-1 | Complete | 895 / 163 | XM_002459533 | No |
| S. bicolor | Poaceae | SbHMA3-2 | Complete | 933 / 201 | XM_002459534 | No |
| S. lycopersicum | Solanaceae | SlHMA1 | Complete | 822 / 39 | AEKE02000758 | Yes |
| S. lycopersicum | Solanaceae | SlHMA-A | Complete | 1196 / 494 | AEKE02004109 | Yes |
| S. tuberosum | Solanaceae | StHMA1 | Complete | 818 / 49 | AEWC01002891 | Yes |
| S. tuberosum | Solanaceae | StHMA-A | Complete | 1192 / 491 | AEWC01030938 | Yes |
| T. aestivum | Poaceae | TaHMA2 | Complete | 1028 / 322 | DQ490135 | No |
| T. cacao | Malvaceae | TcHMA1 | Complete | 813 / 39 | CACC01021667 | Yes |
| T. cacao | Malvaceae | TcHMA-A | Partial | 371 | CACC01013866 | Yes |
| T. halophila | Brassicaceae | ThHMA1 | Partial | 553 / 37 | AK352518 | Yes |
| V. vinifera | Vitaceae | VvHMA1 | Complete | 829 / 40 | XM_002278513 | No |
| V. vinifera | Vitaceae | VvHMA-A | Complete | 986 / 294 | AM454465 | Yes |
| Z. mays | Poaceae | ZmHMA1 | Complete | 823 / 40 | AC199640 | Yes |
| Z. mays | Poaceae | ZmHMA2 | Complete | 1099 / 397 | AC192236 | Yes |
| Z. mays | Poaceae | ZmHMA3-1 | Complete | 898 / 179 | AC190905 | Yes |
| Z. mays | Poaceae | ZmHMA3-2 | Complete | 894 / 180 | AC190905 | Yes |
| Z. mays | Poaceae | ZmHMA3-3 | Complete | 883 / 161 | AC205008 | Yes |
2.2 Annotation and phylogenetic analyses of the heavy metal P1B-ATPase sequences
To deduce the protein sequences from the raw nucleotide sequences, annotation was performed mainly through similarity analyses. The query DNA sequences were compared: (i) to protein sequences translated from validated HMA encoding cDNAs in pairwise comparisons using TBLASTN; as well as (ii) to genome sequences from other species using TBLASTX. The precise positioning of the introns was performed through the recognition of the GT and AG motifs delimiting the 5’ and 3’ ends of introns, respectively. This approach proved to be more precise and reliable than using annotation softwares such as FGENESH, Eukaryotic GeneMark, EuGène or SpliceMachine.
To determine the length of the C-terminal cytoplasmic domain, we started from the analysis performed for the AhHMA4 gene [12], which indicated the position of the transmembrane helices. Using both a sequence similarity approach and the TMHMM software (http://www.cbs.dtu.dk/services/TMHMM/), we determined the position of the last transmembrane domain in all the HMA sequences. We then considered that the C-terminal loop started at the first amino acid located downstream the last transmembrane domain.
For the phylogenetic analyses, full-length amino acid sequences were aligned by CLUSTALW and imported into the Molecular Evolutionary Genetics Analysis (MEGA) package version 4 (http://www.megasoftware.net [24]). All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). Phylogenetic analyses were carried out using the Neighbor-Joining method [25], the Minimum Evolution method [26], the UPGMA method [27], the Maximum Likelihood method [28] and the Maximum Parsimony method [29], along with statistical bootstrapping procedure involving 1000 replicates.
3 Results
3.1 Establishment of a complete set of sequences encoding Zn/Cd/Pb/Co P1B-ATPases
In order to perform the phylogenetic analysis of the Zn/Cd/Pb/Co P1B-ATPases family, it was decided to establish a complete set of sequences. The A. thaliana HMA1, HMA2, HMA3 and HMA4 protein sequences – which were deduced from the nucleotide sequences whose accession numbers were NM_119890, AY434728, AY055217 and AF412407, respectively – were used as starting queries to search for nucleotide sequences encoding similar proteins in the public sequence databases using the TBLASTN program. All the different databases gathering genome sequences were analysed. The ‘expressed sequence tag’ (est) database was not considered because it gathers sequences that are both insufficiently accurate and too short (< 800 bp) compared to the length of the HMA genes (> 2200 bp). Among the retrieved sequences, sequences encoding proteins showing orthology to either of AtHMA1, AtHMA2, AtHMA3 or AtHMA4 according to phylogenetic analyses were added to the set of sequences and sequences showing low similarity with our query sequences were discarded. We primarily considered sequence entries corresponding to entire HMA sequences. When an entry only corresponded to a partial HMA sequence, we did not try to establish a full length HMA sequence by assembling that partial sequence with any another partial HMA sequence present in the databases. This was done to avoid assembling overlapping sequence that would actually correspond to different genes. In order to be sure to establish a complete dataset, the identified sequences were translated into protein sequences that were used again as starting query sequences to search for new similar nucleotide sequences in databases, as described above. Only new sequences encoding proteins showing orthology to either of AtHMA1, AtHMA2, AtHMA3 or AtHMA4 were then added to the original set of sequences. This process was repeated until no additional sequence could be added to the set of sequences, considering the sequences available in the databases at the end of April 2011. Some genomic sequences were not considered, either because they were still too fragmented (this was for instance the case for the Lotus japonicus sequences or for the Phoenix dactylifera orthologue to the AtHMA1 sequence) or because they still harboured a great proportion of ambiguous nucleotides (this was, for instance, the situation for the malusXdomesticus sequences).
An important issue was to establish a dataset devoid of duplicates. Sequences were considered to correspond to potential duplicates (as for instance different alleles corresponding to a same locus) on the basis of high sequence identity (> 98%) in the non-coding regions neighbouring the HMA exons. Potential duplicates were removed from the dataset. However, when two or more HMA sequences, showing very high similarity both in coding and non-coding regions were associated in tandem in a single genomic sequence, they were considered as corresponding to distinct genes. This was for instance the case for the three A. halleri HMA4 tandem duplicates, as shown previously [13]. Because of these different filtering steps, sequences reported in Table 1 can be considered as corresponding to distinct paralogues.
A total of 96 HMA sequences were collected (Table 1 and Supplementary Table S1). The dataset comprised sequences from 32 plant species belonging to 15 botanical families (Arecaceae, Asteraceae, Brassicaceae, Caricaceae, Cucurbitaceae, Euphorbiaceae, Fabaceae, Malvaceae, Nicotianeae, Pinaceae, Poaceae, Rosaceae, Salicaceae, Solanaceae and Vitaceae).
3.2 Naming of the HMA sequences
One step of the work has been to give names to the HMA protein sequences that were identified. The names started with two letters indicating the plant species from which the sequence originated. Then the three ‘HMA’ letters were used to indicate the gene family. Finally the names ended with either figures (1, 2, 3, 4) or letters (A, B, C, D). We kept their names to the sequences that had already been named. This was the situation for the Arabidopsis, the rice, the N. caerulescens and one of the poplar sequences [1,14,30]. For the previously unnamed sequences, we determined the terminal part of the names after completion of the complete phylogenetic analysis described below. When a clear orthology relationship associated the sequence of a previously unnamed heavy metal ATPase to the sequence of an already named heavy metal ATPase, we put the terminal part of the known sequence at the end of the still unnamed sequence. For instance, the Sorghum bicolor HMA protein sequence extracted from accession number XM_002438908 showed a clear orthology relationship to the OsHMA2 sequence; it was thus named SbHMA2 (Table 1 and Fig. 1). Sometimes two or more previously unnamed sequences showed a clear orthology relationship with one specific already named heavy metal ATPase. For instance, the S. bicolor HMA proteins extracted from accession numbers XM_002459533 and XM_002459534 both showed a clear orthology relationship to the OsHMA3 sequence (Fig. 1). These proteins were then named SbHMA3-1 and SbHMA3-2. When no obvious orthology relationship associated sequences encoding previously unnamed heavy metal ATPases to sequences of already named heavy metal ATPases, the -A, -B, -C or -D letters were appended at the end of the sequence names (for instance, GmHMA-A). As detailed below, this applied to all the HMA sequences corresponding to plant species belonging to other botanical families than Poaceae or Brassicaceae (Fig. 1).
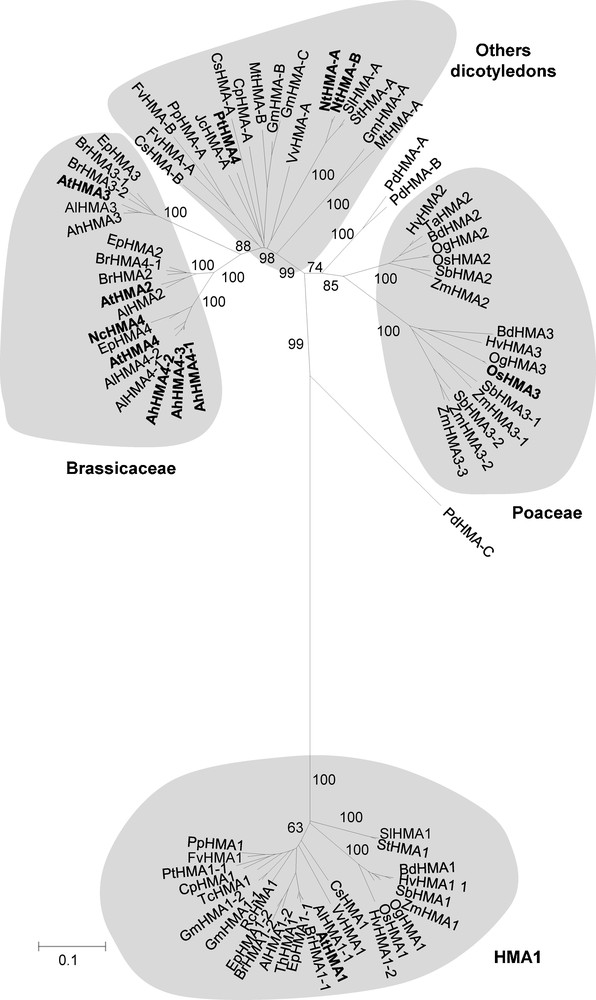
Phylogenetic analysis of the plant Zn/Cd/Pb/Co P1B-ATPase family. The phylogenetic analysis was performed using 83 sequences corresponding to all the proteins that are orthologous to any of AtHMA1, AtHMA2, AtHMA3 or AtHMA4, except those which sequence was less than 550 amino acids long. Different methods were used; they gave similar results. The presented tree was the optimal tree obtained using the Neighbor-Joining method and corresponds to a bootstrap consensus tree inferred from 1000 replicates. The numbers at the branches are confidence values based on Felsenstein's bootstrap method [34]. The tree is drawn to scale, with branch lengths corresponding to the number of amino acid substitutions per site. The sum of branch length is 6.53. All positions containing gaps and missing data were eliminated, leaving a total of 319 positions in the dataset. The evolutionary distances used to infer the phylogenetic tree were computed using the Poisson correction method [35]. Proteins for which a functional role at the plant level has been ascribed are indicated in bold.
3.3 Validation and analysis of the HMA sequences showing orthology to either of the AtHMA1, AtHMA2, AtHMA3 or AtHMA4 sequences
Less than one third of the HMA nucleotide sequences identified as described above corresponded to experimentally validated full-length cDNA clones. The validated sequences were the Arabidopsis and rice ones, as well as a couple of sequences from other plant species such as N. caerulescens, maize, poplar,… (Table 1). Most of the other HMA sequences were collected from on-going genome sequencing projects. They were thus either not annotated or not properly annotated. We thus made the annotation ourselves to obtain the appropriate deduced protein sequences that are given in Supplementary Table S1. In the course of our annotation, we observed that at least four HMA sequences most likely corresponded to pseudogenes. Indeed, these sequences harboured STOP codons within the coding sequence (BrHMA4-2 and GmHMA-D) or displayed only few of the exons (GmHMA-D, PtHMA-B and VvHMA-B). However, in some instances, the genome sequencing was not finished, or the authors who deposited the sequences in the databases only performed partial sequencing. In these situations, we did not consider the incomplete HMA sequences to correspond to pseudogenes and we quoted them as ‘partial’ in Table 1.
From the analysis of the validated set of sequences, we observed that the number of HMA paralogues varied depending on the species considered (Table 1). For instance, most of the species possessed only one HMA1 copy; only Arabidopsis lyrata, barley (Hordeum vulgare) Eutrema parvulum, mustard (Brassica rapa) and soybean (Glycine max) displayed two complete copies. If we now focus on the orthologues of AtHMA2, AtHMA3 or AtHMA4, the situation is more complex. Rice and the strong-spined medick (Medicago truncatula), which genomes are fully sequenced, possess only two paralogues in their genome, while mustard, whose genome is still incompletely sequenced, already displays five paralogues (Table 1). Within the same botanical family, different species display different numbers of HMA copies (compare Z. mays or sorghum to rice for instance, or A. halleri and A. lyrata to A. thaliana in Table 1).
The protein structure of the different HMAs was also highly variable. If we concentrate on the Zn/Cd/Pb/Co P1B-ATPases other than the HMA1 orthologues for which a full length protein sequence is available, performing a multiple sequence alignment showed that only the first ∼700 amino-acids of the proteins, which harbour the transmembrane domain, display a high level of similarity (data not shown). The cytoplasmic C terminal parts of the proteins appeared to be completely different between orthologues, as well as between paralogues. In particular, they display markedly different lengths between the orthologues, comprising from 58 to 703 amino acids, the average length being ∼290 amino acids (Table 1 and Fig. 2). Intriguingly, some of the Brassicaceae HMAs, the HMA3 ones, display a specific characteristic: their C-terminal part is only ∼60 amino acid long while the C-terminal parts of all the other orthologues to AtHMA2, AtHMA3 or AtHMA4 are much longer (Table 1 and Fig. 2).
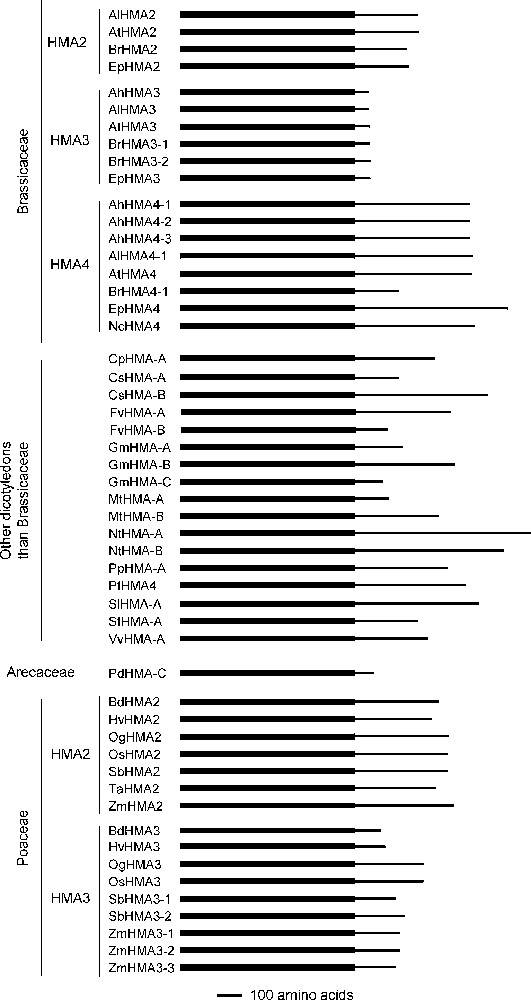
Structural comparison of the complete proteins corresponding to orthologues to AtHMA2, AtHMA3 and AtHMA4. The simplified protein structure is presented for the 54 proteins that are orthologous to AtHMA2, AtHMA3 and AtHMA4 and for which a complete sequence is available. The thick and thin portions of the lines symbolise the N-terminal transmembrane domains and the cytosolic C-terminal domains of the proteins, respectively. Proteins are ordered in agreement with the phylogenetic analysis presented in Fig. 1.
3.4 Phylogenetic analyses
A phylogenetic analysis was performed considering the 83 collected HMA proteins for which sequences longer than 550 amino acids were available (Fig. 1). The C-terminal region of the protein could not be considered in this analysis since no sufficient similarity could be detected between the different homologues. Different methods were used including maximum likelihood-, distance- and parsimony-based ones. They all produced nearly identical trees. The very few observed permutations concerned terminal branches of the trees; these permutations did not change the overall organization of the trees (data not shown). This convergence between the results from the different methods warrants the robustness of the phylogenetic analysis.
The phylogenetic analysis revealed that the orthologues of AtHMA1 were all grouped with each other in one clade, whatever their species of origin (Fig. 1). In contrast, the phylogenetic relationship linking the orthologues of AtHMA2, AtHMA3 or AtHMA4 was unusual: the structure of the phylogenetic tree made from these orthologues was strongly dependent from the botanical family to which the sequences could be related to rather than from the different protein isoforms (Fig. 1). Three subgroups were revealed. All the orthologues of AtHMA2, AtHMA3 or AtHMA4 that were identified in Poaceae species constituted a first specific subgroup, within which two separate clades grouping orthologues of OsHMA2 from rice on one side and orthologues of OsHMA3 from rice on the other side could be distinguished. The orthologues of AtHMA2, AtHMA3 or AtHMA4 that were identified in dicotyledonous species were split into two subgroups. One of these subgroups comprised HMAs identified in Brassicaceae species only, and the other one comprised HMAs identified in all the other dicotyledonous species. Within the Brassicaceae subgroup, the HMAs were arranged into three different well-separated clades, one of which contained AtHMA2, the second of which contained AtHMA3 and the third of which contained AtHMA4. In contrast, there was no clear arrangement of the orthologues of AtHMA2, AtHMA3 or AtHMA4 that were identified in non-Brassicaceae dicotyledonous species (Fig. 1); thus these orthologues could not be directly related to either of AtHMA2, AtHMA3, AtHMA4, OsHMA2 or OsHMA3. Finally, three sequences extracted from the monocotyledon P. dactylifera species appear to be scattered in-between the Poaceae sequences and the dicotyledonous sequences.
4 Discussion
The Zn/Cd/Pb/Co P1B-ATPases have been extensively studied in Brassicaceae species such as A. thaliana [7–11,20], A. halleri [13] or to a lesser extent N. caerulescens [14,17]. Recent reports also described the function of the rice OsHMA3 sequence [18,19]. These heavy metal ATPases play a critical role in controlling the zinc or cadmium translocation from roots to shoots or in controlling the transport of cadmium or lead from the cytoplasm into the vacuole. In Brassicaceae, different paralogues are specifically responsible for each of these two functions. HMA2 and HMA4 are involved in the control of the zinc and cadmium translocation from roots to shoots through their ability to transport zinc or cadmium into the xylem [9–11], while HMA3 is responsible for the storage of cadmium or lead into cell vacuoles and does not control the root to shoot translocation of metals [20]. Amazingly, the situation seems to be different in Poaceae. In rice OsHMA3 controls cadmium translocation from roots to shoots while being involved in the storage of cadmium into root cell vacuoles [18,19]. Up to now, the causes of these functional differences are still unknown. One hypothesis could have been to correlate differences between the in planta roles of AtHMA2, AtHMA3, AtHMA4 and OsHMA3 with differences in protein structure. It, however, appears still premature to do so. A major difference between the HMAs is the length of the cytosolic C-terminal region. While AtHMA3 and its Brassicaceae orthologues display a very short C-terminal region, the other orthologues of either of AtHMA2, AtHMA3 or AtHMA4 harbour a long one that possesses many histidines as well as cystein doublets. The AtHMA2 C-terminal region was proposed to chelate Zn2+ and to increase the protein turnover and enzyme velocity [22], but this knowledge does not help to infer the functional differences between the HMAs. The AtHMA2 C-terminal region was also proposed to play a role in the subcellular targeting of the protein [23], but so far, little is known about the sorting motifs involved in the targeting to different membranes and again, this does not help to infer the functional differences between the HMAs.
The above-mentioned knowledge acquired from the study of AtHMA2, AtHMA3 or AtHMA4 is implicitly considered to be transferable to orthologous HMAs in other species. However, such a transfer may not be straightforward. The present phylogenetic analysis supports this latter proposition. First of all, we observed that the number of HMA copies showing orthology to either of the AtHMA2, AtHMA3 or AtHMA4 paralogues was markedly variable, spanning from two to at least five depending on the species considered. In addition, the between-species variation in the copy number of these HMAs was great even within a same botanical family. This indicates that as far as the Zn/Cd/Pb/Co P1B-ATPases gene family is concerned, a great number of duplication events occurred independently and recently, i.e. after speciation. For instance, three copies of the sole HMA4 gene are present in A. halleri while only one copy is present in A. thaliana, which diverged from A. halleri less than 15 million years ago [13]. In addition, we observed a major variation in the length (from 58 to 703 amino acids) of the cytosolic regulatory C-terminal domain between paralogues within a species and more importantly between orthologues across species. These observations indicate that the Zn/Cd/Pb/Co P1B-ATPases gene family undergoes a very dynamic evolutionary process. This is for sure a first reason why uncovering orthology relationships within this family is challenging.
The present phylogenetic analysis also revealed that no strict orthology relationship links the monocotyledon copies of the Zn/Cd/Pb/Co P1B-ATPases to the dicotyledon ones. Indeed, the two HMA2 and HMA3 subfamilies gathering the Zn/Cd/Pb/Co P1B-ATPases from Poaceae displayed greater similarities with each other than with any of the HMAs identified from the other monocotyledon species P. dactylifera or from any of the dicotyledons. The same observation could be made concerning the dicotyledon members of this family themselves. Indeed, no strict orthology relationship links the Brassicaceae representatives to the non-Brassicaceae ones. Altogether, these observations indicate that uncovering orthology relationships within the Zn/Cd/Pb/Co P1B-ATPase family is probably not possible when phylogenetically distant species are examined.
Although there are exceptions to the rule it is assumed that orthologues share similar functions while paralogues more likely display different functions [31]. Though, since no clear orthology relationship can be established between Zn/Cd/Pb/Co P1B-ATPases from Brassicaceae and Zn/Cd/Pb/Co P1B-ATPases from other plant species, is the functional diversity that is observed between these ATPases in A. thaliana (HMA2 and HMA4 vs. HMA3) also encountered in non-Brassicaceae species? A recent patent deposition describing RNAi inactivation of tobacco Zn/Cd/Pb/Co P1B-ATPases showed that inactivation of these genes resulted in the complete inhibition of the translocation of cadmium from roots to shoots [32]. This suggests that the tobacco Zn/Cd/Pb/Co P1B-ATPases display a similar function as the A. thaliana HMA2 and HMA4, but not as the A. thaliana HMA3. In rice, the situation seems to be more contrasted. As already mentioned, OsHMA3 plays both a similar role as AtHMA3 at the cell level and an opposite role compared to AtHMA2 or AtHMA4 at the whole-plant level [18,19]. In addition, OsHMA3 seems to be able to transport cadmium but not zinc, which discriminates it from AtHMA2 and AtHMA4. Thus OsHMA3 plays an overall specific and different role compared to AtHMA2, AtHMA3 or AtHMA4. These analyses lead to the following working hypotheses that Zn/Cd/Pb/Co P1B-ATPases might play specific roles depending on the species considered, that the functional diversity observed for the different A. thaliana AtHMA2, AtHMA3 and AtHMA4 paralogues could be specific to the Brassicaceae species, and thus that the AtHMA3 function might be unique to the Brassicaceae species. In this respect, since AtHMA3 was shown to contribute to Zn2+, Cd2+, Pb2+ and Co2+ tolerance [20], Brassicaceae species might be best suited to support the corresponding heavy metal constraints and thus to be used in phytoremediation approaches aiming at cleaning Zn- or Cd-polluted soils. Actually, most of the Zn- and Cd-hyperaccumulating species belong to this botanical family [33].
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
W.Z. was supported by a scholarship from the Tunisian Ministry of Higher Education and Scientific Research (LR10CBBC02), and then by a scholarship from the “Agence universitaire de la francophonie” (AUF). We thank Dr F. Gosti and Dr L. Marquès for critical reading of the manuscript, and A. Adiveze, H. Afonso, C. Baracco, H. Baudot, F. Bourgeois, C. Dasen, X. Dumont, C. Fizames, J. Garcia, S. Gélin, F. Lecocq, V. Papy, V. Rafin, G. Ruiz and C. Zicler, for technical and administrative supports.


