1 Introduction
Elliptic Fourier descriptor analysis is a method for contour description. A closed curve is a continuous periodic function of a parameter, so it can be alternatively represented as a sum of cosine and sine functions of growing frequencies, affected by coefficients referred to as ‘Fourier descriptors’ (FDs). The sum of these cosine and sine functions converges towards the initial contour, as the number of harmonics increases. Each harmonic is actually an ellipse, completely defined by its period and its FDs.
Elliptical Fourier descriptors were first described in the two-dimensional space [1–4] with some variety regarding the variable used for parametrization (arc length, angle, etc.). A plane, closed contour is expressed as a sum of two-dimensional ellipses of decreasing areas. The FDs can be used as morphometric variables in multivariate analysis, allowing for the distinction of groups within a set of shapes. Most applications in two dimensions are related to biological issues [5], for anthropology [6–8], anatomy [9], evaluation of the response to orthodontics treatment [10], etc. Hand-written character recognition [2,11,12] or aircraft recognition algorithms [3,13] are other areas of use. The FDs allow for the computation of invariants of the contour, independent under translation, rotation or change in the starting point on the contour [2,4]. In addition to providing interesting information compression, FDs can be used for smoothing contours, by keeping only the first harmonics.
In three dimensions, two different problems may be addressed. One is the study of bounding surfaces [14–19] where shapes are described as sum of ellipsoids. The other is the study of closed lines embedded in the three-dimensional space. Few papers have been published on the subject. Numerical invariants of three-dimensional contours are computed in reference [20] to study the influence of noise and distortion for shape recognition purposes. In reference [21], lines are simplified as ‘chaincodes’, reducing the complexity of the initial contour. FDs are computed using this formalism, and normalized to make them independent of scale change and orientation of the global contour. In reference [22], the author makes the FDs independent of the starting point and uses them in the cardiology field to identify supraventricular beats by analyzing the 3D curves formed by the electrocardiogram vector (QRS loop). In reference [23], the authors used 3D FDs to optimize the fitting of protection harnesses to body shape. Some works make use of the results in two dimensions for studying surfaces, either by ‘slicing’ the initial 3D surface [24–26], by combining projections [27] or using invariants [28]. None of these papers give an explicit correspondance between FDs and their geometric parameters.
In this article, we study lines embedded in three-dimensional space. Real world objects, more particularly, anatomical structures, tend to be three-dimensional rather than two-dimensional. Tomographic imaging such as scanner and MRI, followed by appropriate image processing, may be used to create three-dimensional objects. We focused on lines rather than surfaces for several reasons. First, mathematical simplicity: Lines may be parametrized by one-variable functions while surfaces require the handling of multiple variable functions. Secondly, many anatomical structures (such as skull or heart) are not closed surfaces, making irrelevant the use of global surface descriptors in the vicinity of the boundary. Unlike reference [21], we did not restrict ourselves to chaincodes, but rather dealt with arbitrary 3D lines.
We start from two 3D lines. Each line represents a contour and has a set of coordinates (sampling) describing it. The aim of this article is to address the following question: how can we compare the two contours? When the sampling of the points on the contours is done in two different Cartesian coordinate systems, we cannot compare directly the lines but we need a transformation step. Our approach is to use the decomposition as elliptical Fourier descriptors to find, for each object, an intrinsic Cartesian coordinate system to perform this transformation. This system will be specific to each contour. As stated earlier, each harmonic of the decomposition of a contour is an ellipse defined by its period and FDs. The first harmonic is an ellipse roughly approximating the contour. We use it as the reference plane of the new Cartesian coordinate system, its symmetry axes being the two axis lines X, Y. We also use the area of this first harmonic to account for the potential scale difference in the two coordinate systems. Making ue of these results, we compute an algorithm for rescaling and reorientation of 3D objects, by normalizing their FDs or their coordinates on the first harmonic.
An ellipse can be described in two ways: either by giving the coefficients of its parametric equation (six numbers in three dimensions), or by giving the geometrical parameters: semi-axis lengths, angles for the orientation (three numbers in three dimensions), starting point on the contour and direction of motion when the parameter increases. In order to perform the normalization-reorientation steps described above, we must know the equations linking the geometrical parameters of a given ellipse (here, the first elliptical descriptor) to the coefficients of its parametric equation (the FDs of the first harmonic). This has been addressed by reference [3] in two dimensions, but not in three dimensions. This is the subject of the Section 2.
To introduce the formalism later used in this article, let us consider a closed curve M(t) of coordinates (x(t), y(t), z(t)). It can be described as a sum of ellipses using the Fourier series expansion of its coordinates:
So
2 Relations between Fourier coefficients and geometrical parameters for ellipses in the three-dimensional space
To determine the relations between the coefficients (xc, xs, yc, zc, zs) and the geometrical parameters a, b, α, β, γ, φ, we start from an ellipse in the two-dimensional plane, defined by its axes and , its period T, a phase shift φ (corresponding to the position of M at t = 0). Its parametric equation is given by:
Let us define now the rotation of this ellipse in the three-dimensional space for any given orientation. For this purpose, we use Euler angles α, β, γ, illustrated in Fig. 1. Any orientation can be achieved by composing three elemental rotations rα, rβ, rγ.
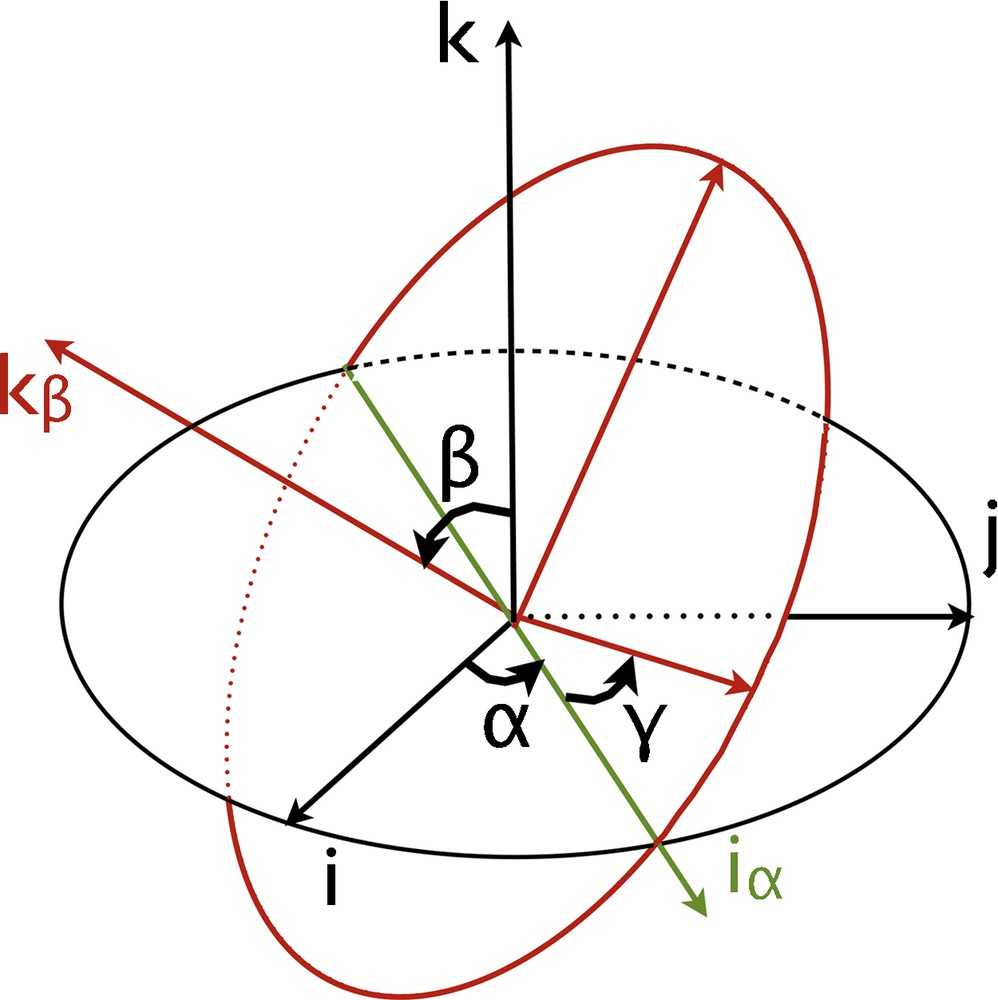
Illustration of the Euler angles used in this study.
Here, rα is a rotation of angle α about the axis, whose matrix Rα is written in the basis ; rβ is a rotation of angle β about the axis, with , whose matrix is Rβ; rγ is a rotation of angle γ about the axis, with , whose matrix is Rγ.
The matrix Ω of this transformation ω, written in the basis , is:
The (Ωij)i, j = 1,…, 3 satisfy:
| (1) |
| (2) |
| (3) |
So any ellipse whose parametric equation is (X(t), Y(t), Z(t)), can be written as
The problem is equivalent to finding a, b, α, β, γ, φ, given xc, xs, yc, ys, zc, zs, bound by the following equations:
and the same for Sy and Sz by replacing Ω1i by Ω2i and Ω3i, respectively.
Using Sx, Sy and Sz in Eqs. (1) to (3), we get:
| (4) |
| (5) |
| (6) |
Eq. (4) has four solutions φ1,…,φ4:
Each φi gives two different solutions for a and b, one positive and one negative.
From the definition of Ω,
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
Using Eqs. (1) to (3), Eqs. (7) and (8) are redundant. Sx, Sy and Sz give Ωij as functions of φ, a, b. So:
Let us define:
- • If w > 0, the solutions for (α, β) are:
(α*, β*)
and (α* + π, −β*)
- • If w < 0, solutions are:
Eqs. (11) and (12) unambiguously define γ, knowing a, b, φ, α, β.
Any given value for φ gives two possible values for a and for b. Any given value of the triplet (φ, a, b) gives two possible values for the triplet (α, β, γ). The set of solutions has then 32 elements, which reflects the symmetry of the problem. For computer programming purposes, one can impose conditions to get a unique solution:
1. φ ∈] − π/4, π/4[
2.
3.
which gives:
3 Normalization of Fourier descriptors and coordinates
In order to address our initial problem of contour comparison, we make use of the preceding results. For two contours, we can compare either their Fourier descriptors, or their coordinates. In either case, we want to transform these numbers, to write them in a new system of coordinates, intrinsic to the contour. As stated earlier, this is achieved by using the first harmonic. This lets us create a rescaling – reorientation algorithm, a required step before contour comparison.
3.1 Normalization of Fourier descriptors
The equations stated earlier give the necessary tools to transform the Fourier coefficients of the k-th harmonic , (or equally α, β, γ, φk, ak, bk, αk, βk, γk) to make them independent of the global size of the contour (normalization), of the orientation (reorientation), of the starting point on the contour (phase shift correction) and of the direction of motion on it. We use the first harmonic, in a similar approach to [21].
3.1.1 Rescaling
We normalize the coefficients so that the area of the first harmonic will be equal to 1. The area of the first harmonic is given by:
3.1.2 Reorientation
We change the orientation of the contour so that the axes of the first harmonic be aligned with X and Y. It can be easily achieved through inverting the transformation ω described in Section 2. For the first harmonic, the angles α1, β1, γ1 define ω1. Let be the Fourier coefficients of the k-th ellipse, after the reorientation step. We have:
3.1.3 Phase shift correction
The departure point on the ellipse has no morphologic relevance. What operation needs to be done to the first harmonic to have a zero phase shift at t = 0? Let be the mobile point describing the first harmonic after correction of the phase shift.
so
The same transformation will hold true for each harmonic, :
3.1.4 Direction of motion
Fourier coefficients of two similar ellipses with opposite direction of motion of the point M (t) across time will be bound by the following equations:
We must impose the same change in the direction of motion to all harmonics. For instance, one can test the sign of (which is the coefficient after reorientation); if negative, replace all the by . Imposing is equivalent to a direct sense of motion of the first harmonic (after the reorientation step described above).
Combining all the preceding steps, we obtain a global formula for the new Fourier coefficients :
If is negative, the new coefficients will be:
3.2 Rescaling and reorientation of the initial coordinates
For comparison purposes, the only relevant steps are normalization and/or reorientation. We also subtract the coordinates of M0, the center of gravity, to correct the translation of the origin of the Cartesian coordinate system. Neither the shift of the starting point nor the direction in which the points are sampled are of interest. Let (xp, yp, zp) be the coordinates of the p-th point on the initial contour. After normalization, reorientation and translation to the origin of the system, the final coordinates will be:
| (13) |
4 Application examples
4.1 An overview of the use of the proposed algorithm for re-orientation and re-scaling applied to a patient X-ray CT
Fig. 2 shows an example of how our algorithm can be used for a three-dimensional re-scaling and re-orientation of two CT imaging exams obtained in different size and position. Such a technique could be used for precise and automated comparison, for a given patient, of images between several investigations (e.g., assessment of tumor size response to treatment). A patient abdominal CT scan was performed (Toshiba 64 slices, American Medical System, Tustin, CA, USA) and the axial slices were processed using a volume rendering software to display the pelvic bone structures (workstation ADW, GE Healthcare, Waukesha, USA). The three-dimensional line of the pelvic outlet contour was manually sampled leading to 43 points which coordinates were given by the imaging software (Figs. 2a, c). We then simulated a different position and a size reduction of the patient pelvis by mathematically changing the coordinates of the initial points by a rotation and scale matrix (Fig. 2b) of which a new set of coordinates of the pelvic outlet was obtained as illustrated in Fig. 2d. We then applied our algorithm to normalize and reorient the two different pelvic outlets, their corresponding first harmonic being the reference (X, Y) plane. Re-orientation and normalization steps of the algorithm give the same contour illustrated in Fig. 2e, whatever set of coordinates is used as an input. We then represented the new pelvic outlet within the initial bone structure (Fig. 2f). This example shows how the algorithm presented here can be used to reorient and normalize three-dimensional contours from different CT imaging acquisitions to perform a comparison between them.
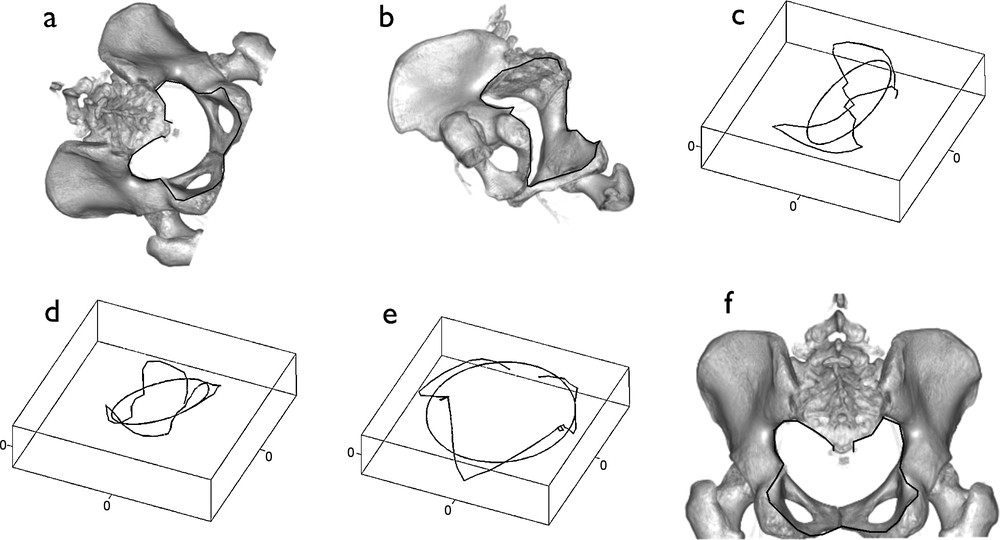
Overview of the algorithm. (a) and (b) show two CT scan bone volume rendering on which the pelvic outlet has been outlined (black line). (b) is a mathematically rotated and rescaled image of (a). Corresponding pelvic outlets alone and first elliptical Fourier descriptor are shown respectively in (c) and (d). Notice that orientation and area of these ellipses are different for the two images. The normalization-reorientation steps of the algorithm are then performed on the two contours, by making the first harmonic the reference plane of the contour and by normalizing the distances in order to set the area of the first harmonic equal to 1. The same contour, illustrated in (e), is obtained after the normalization-reorientation steps of the algorithm, whether starting from the (c) or (d) contours. Finally, the new pelvic outlet is represented in (f) superimposed to the initial bone structures.
4.2 Morphometrical analysis of the foramen magnum contour in a murine model of X-linked hypohidrotic ectodermal dysplasia
X-linked hypohidrotic ectodermal dysplasia (XLHED) is a genetic disorder due to a mutation of Eda gene [29]. The clinical phenotype of individuals affected by HED is complex and associates hypotrichosis, dental agenesis and morphological defects, onychodysplasia and eccrine glands aplasia or hypoplasia, being the main cardinal features [30]. Craniofacial and anthropometric changes are also described in XLHED and consist in reduced and retrognatic maxilla, frontal prominence, cranial base modifications, reduced facial convexity and facial height with deficiencies in skeletal sagittal and transversal growth [31–33]. Although the tooth abnormalities in Tabby mutant mice (the murine model of XLHED) have been extensively studied, the characterization of the craniofacial complex has received less attention. Therefore from 3D microCT reconstructions of the skull, the contour of the foramen magnum (a 3D line) was quantified using 3D Elliptical Fourier analysis, allowing quantitative comparisons between Tabby specimens and their wild type controls.
EdaTa mice were generated from the inbred Ta strain B6CBACa Aw-J/A-EdaTa/J-XO (Jackson Laboratory, USA). Mice phenotype was identified according to external morphological criteria: bald patches behind ears and deformities in the distal portion of the tail for hemizygous males (EdaTa/Y), striping of the coat for heterozygous females (EdaTa/ + ). WT mice were generated by inbreeding wild-type animals derived from the B6129PF2/J strain considered as control for strains designated by B6. WT mice and Tabby heterozygous females and homozygous males were genotyped according to previously published protocols [34] through PCR amplification using specific forward and reverse oligonucleotides (3F-7R, 6F-8R and 5F-6R), derived from −3 kb to +1.9 kb murine genomic sequence surrounding exon 1 of the WT Eda gene. The presence of a deletion in Eda gene exon 1 was observed in Ta specimens, as the 6F-8R primers allowed for amplification only in Ta heterozygous females and WT. The expected genotype of each Ta mice was confirmed by the absence of the 5F-6R primed fragments in Ta males. Mice population sample included 24 specimens, seven hemizygous males (EdaTa/Y), six heterozygous females (EdaTa/ + ), six control WT females and five control WT males. All the specimens studied were adults (3-month-old mice). Mice were killed by intraperitoneal lethal injection of pentobarbital. The experimental protocol was designed in compliance with recommendations of the EEC (86/609/CEE) for the care and use of laboratory animals. The animals were fixed in 1:10 37% formol, 3:4 100% ethanol, and 3:20 distilled water solution for 15 days. Imaging of full heads was performed using a microCT system (eXplore CT 120, GE Healthcare, Waukesha, WI, USA). The protocol used acquired 360 views on 360°, at 100 kV, 50.0 mA, with an exposure time of 20 ms. The duration of one acquisition was less than 4 min. A Feldkamp algorithm of backprojection led to the reconstruction of a volume made of cubic voxels of 50 μm × 50 μm × 50 μm. Steps of the image analysis procedure were performed using MicroWiew (GE Healthcare, Waukesha, WI, USA). A 3D virtual reconstruction of the skull was computed using the isosurface volume rendering module with a threshold value set at 600 HU (Fig. 3). Each foramen magnum outline was then manually recorded as a series of 20 sampling points represented by their 3D Cartesian coordinates (Fig. 3). The degree of distinction among mouse strains was then assessed and displayed graphically using discriminant analysis performed with Statistica 7.1 software package (Statsoft Inc., Tulsa, OK, USA). Correlations between 3D elliptical Fourier descriptors and discriminant axes were calculated. The unbiased Mahalanobis D2 distances, corrected for unequal sample sizes between the centroids of each group were also calculated to express inter-mouse strains variability. The discriminant analysis of the elliptical descriptors was limited to the first four harmonics because the multivariate analysis was no more significant after these harmonics (WilksLamda values p ≥ 0.05). The first two discriminant functions (DF1, DF2) described 86.3% of the total variance, the first accounting for 80.9% of the total variance, the second DF for the remaining 5.4%. Illustrated in Fig. 3, hemizygous Ta males and heterozygous Ta females clearly departed from the corresponding WT groups as shown by the significant distances separating the EdaTa/Y and EdaTa/+ specimens from the other mice groups (Table 1). The greatest difference occurred between WT males and females illustrating the sexual dysmorphism. The lower difference occured between Ta females and corresponding WT as demonstrated by the inter-group distances illustrated in Fig. 3 and Table 1.
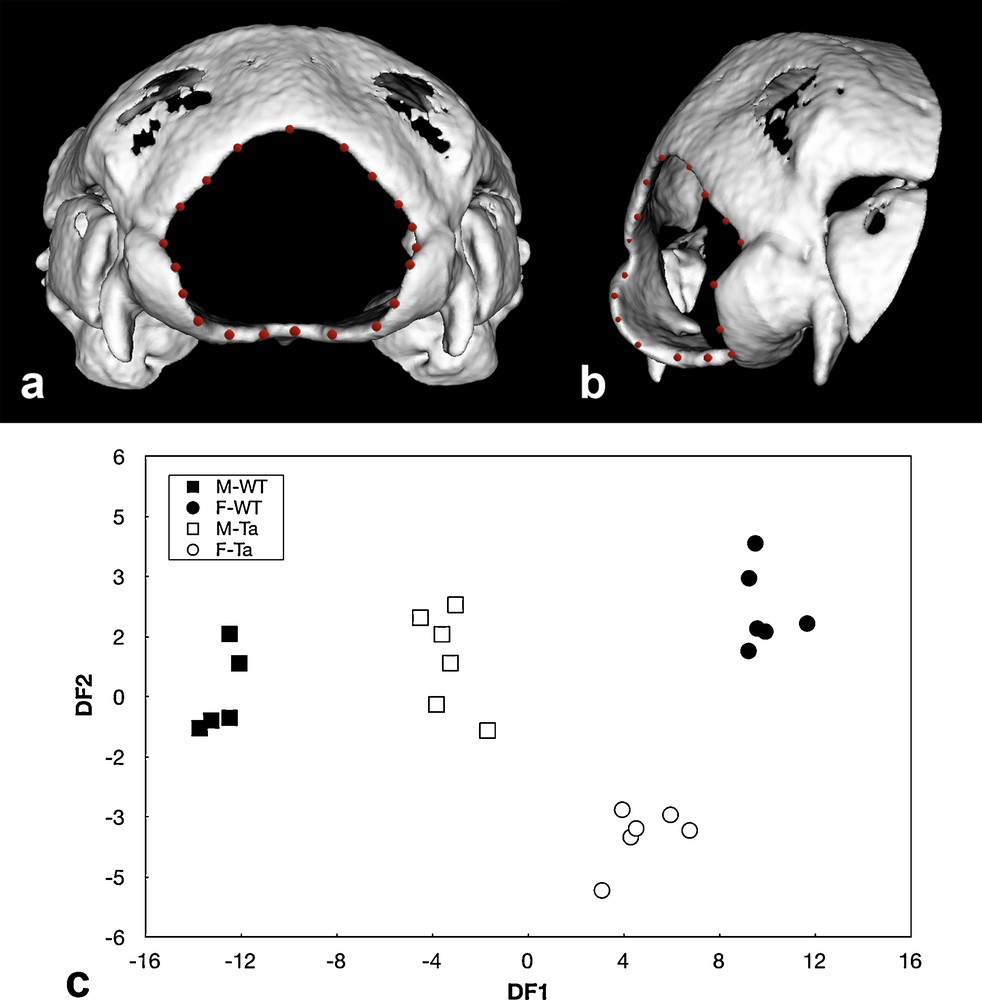
Isosurface 3D microCT reconstructions of wild type and Tabby mice skull. (a) posterior view and (b) postero-lateral view showing also the sampling points of the three-dimensional contour of the foramen magnum outlined in red. (c) discriminant analysis of the 4 first elliptical descriptors of the foramen magnum of the four mice groups (M-WT: wild type males, F-WT: wild type females, M-Ta: Tabby hemizygous males, F-Ta: Tabby heterozygous females).
Unbiased Mahalanobis D2 distances (corrected for unequal sample sizes) between the centroids of each mice group.
| M-WT | F-WT | M-Ta | F-Ta | |
| M-WT | 0 | 518.3 | 114.3 | 326.1 |
| F-WT | 518.3 | 0 | 193.9 | 59.6 |
| M-Ta | 114.3 | 193.9 | 0 | 93.9 |
| F-Ta | 326.1 | 59.6 | 93.9 | 0 |
5 Discussion
The algorithm presented in this article makes use of the elliptical Fourier descriptors to reorient, rescale and compare three-dimensional curves. It makes use of the first harmonic to define an intrinsic Cartesian coordinate system in which Fourier descriptors, or alternatively coordinates, are computed. Potential applications of this algorithm are numerous but we focus the discussion to biological and medical examples. Our first application example demonstrates how the algorithm can be used to reorient and normalize 3D lines of the same pelvic bone anatomical structures after simulation of two different patient size and position from an X-ray CT exam. In biomedical imaging, comparison of sequential three-dimensional contours across time for the follow-up of a particular patient needs such steps of normalization and reorientation. In clinical practice, the algorithm can also be used to rescale, reorient, and compare two exams acquired in two different conditions, such as stress vs. rest in myocardial perfusion single photon emission computed tomography (compared to clinical existing nuclear medicine softwares where this step of reorientation is obtained manually). Alternatively, one may want to describe 3D anatomical lines to assess the feasibility of a certain procedure. For instance, in obstetrics, pelvimetry evaluates the size of pelvic inlet and outlet to know if a vaginal delivery is possible, or if a cesarean section is required. Once clinical, this assessment is now helped with CT-measurements but does not take into account the whole 3D contour information. It is well-known that these measurements have poor predictive value (see for example [35]). Extraction of the 3D line and its subsequent analysis with our algorighm may lead to precious information regarding the possiblity of vaginal delivery. More broadly, compared to manual anatomical landmarks and distances analysis, elliptical Fourier descriptors of bi-dimensional contours of anatomical structures (obtained for instance by X-ray planar projection) have demonstrated a great potential for shape comparison purposes [5–9]. However, elliptical Fourier descriptors of three-dimensional contours are more anatomically relevant, since most anatomical structures are three-dimensional. It can be used to sort animals for taxonomic purposes, or to study gene influence on phenotype as shown in our second example about cranial dysmorphoses in a murine model of XLHED. In this case, the 3D elliptical Fourier descriptors algorithm was applied to a particular 3D line of an open bone structure of the skull (the foramen magnum), allowing for a clear distinction between homozygous and heterozygous Ta mice and their WT controls. This particular example illustrates the interest of the proposed method for open anatomical structures like the foramen magnum whose three-dimensional morphology are generally difficult to analyze. Elliptical Fourier descriptors analysis applied to three-dimensional contours treats the shape as a single entity and allows retrieval of the entire anatomical profile. For better data reproducibility, automated three-dimensional contour detection from biomedical isotropic tomographic images should be preferred to manual extraction. Entire outlines of biological objects contours can be obtained from sets of non invasive digital isotropic tomographic images sources (CT or MRI images for instance). Based on differences in contrasts, many filtering and tresholding data processing softwares are able to trace automatically the 3D boundaries of particular anatomical structures. It avoids the difficulties of localizing specific landmarks, especially for open structures [36,37]. A whole automated and efficient shape analysis procedure could be then achieved starting with an extraction of 3D surface by volume rendering image processing software followed by a selection of singular aligned points on the surface as an input to the algorithm presented in this paper. This procedure could be of particular interest for automated screening in large scale mouse phenotyping programs. Finally, computational time is not a major concern given the speed of the algorithm.
6 Conclusion
The algorithm presented in this article computes, for the first time, the relations between the Fourier coefficients of a contour embedded in the three-dimensional space as a sum of ellipses and its corresponding geometric parameters. These relations are then used to create an algorithm for the rescaling and reorientation of 3D objects, and contour comparison. The usefulness of this algorithm is demonstrated for morphometrical comparison purposes using 3D anatomical profiles obtained from biomedical clinical and preclinical X-ray computed tomography.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
The authors thank P. Choquet, M. Schmittbuhl and R. Godefroy for helpful discussions. A special thank to F. Hubele for his help throughout this work.


