1 Introduction
Epstein-Barr virus (EBV) is a ubiquitous tumor-causing virus, infecting more than 90% of the adult in world population and achieves life-long infection in B lymphocytes asymptotically [1]. Several epithelial cancers are now proven to be related to the oncogenic properties of EBV, including nasopharyngeal carcinoma (NPC), oral, and gastric cancers [1–3]. There are nearly 100 EBV genes in the EBV genome; however, there are only a few genes expressed in latent infection. There is only a limited set of latent genes including six EBV nuclear antigens (EBNA1, -2, -3A, -3B, -3C and LP); three latent membrane proteins (LMP-1, LMP-2A and LMP-2B); two non-coding EBV-encoded RNA (EBER1 and EBER2); and transcripts from the BamHI A region of EBV genome. Latent genes have been shown to have significant transformation ability in EBV infected B-cells [4].
The Epstein-Barr virus latent membrane protein-1 (LMP-1) is an integral membrane protein with a molecular weight of approximately 63 kDa [5]. LMP-1 expression in nude mice results in B-cell lymphoma [6]. LMP-1 induces a signaling response in cells that mimics a constitutively active form of the B-cell-surface molecule CD40 [7].
Latent membrane protein 2 is divided into two subtypes, LMP-2A and LMP-2B or TP-1 and TP-2 [8]. The unique N-terminal of LMP-2A consists of a 119 amino acids hydrophilic cytoplasmic domain and is mainly responsible for LMP-2A functions [9]. N-terminal of LMP-2A contains an immunoreceptor tyrosine-based activation motif (ITAM) that modulates signal transduction of the B-cell receptor (BCR) and the T-cell receptor (TCR) [10]. However, the function of LMP-2B is mainly unknown.
EBV-encoded LMP-1, LMP-2A and LMP-2B are frequently co-expressed in many EBV-associated malignancies such as NPC, Hodgkin's disease and immunoblastic lymphoma [11–14]. These proteins share some similarities in protein structure and involvement in regulating signaling pathways. Since these proteins share a common identity, they may co-localize to a same cellular compartment [15–17]. Due to their co-localization and pathway inducing abilities, interactions between these proteins may exist. Studies show that LMP-2B modulates the activity of LMP-2A by preventing it from phosphorylation [17] and LMP-2A augments the signaling of LMP-1 through extending LMP-1 turnover [18].
LMP-1 is a well-known classical oncoprotein and proven to transform rodent fibroblast and promote tumorigenesis [19]. On the contrary, studies on LMP-1 suggest that overexpression of LMP-1 exhibits toxicity to the cells [20]. LMP-1 induces type II ligand independent autoactivation of CD95/Fas with Caspase-8 mediated apoptosis [21]. This is thought to cause the cytotoxicity effect of LMP-1. On the other hand, LMP-1 may interrupt cell proliferation when introduced into B-cell lines and NPC cell lines [22–24]. Induction of LMP-1 expression completely inhibits cell proliferation for 4–5 days and is most probably due to the induction of G2 or M phase cell cycle arrest [25]. However, the cytostatic effect of LMP-1 which was due to the arrest of cells at the G2/M phase is poorly understood.
Recently, research in 14-3-3 proteins has achieved increasing interest in many fields of biology. 14-3-3σ protein belongs to a highly conserved acidic protein family which has seven isoforms in mammals. Most of the 14-3-3 family members are involved in regulating signaling pathway, apoptosis, adhesion, cellular proliferation, diferentiation and survival [26]. Of all the seven isoforms, only 14-3-3σ seemed to be involved directly to human cancers [27–29]. 14-3-3 sigma is expressed mostly in epithelial cells and has been associated with cancers including breast, nasopharyngeal, endometrial adenocarcinoma, prostate, epithelial ovarian, and colorectal [27,28]. 14-3-3σ is a p53 regulated inhibitor of G2/M progression [26]. Ectopic expression of 14-3-3σ protein results in G2 arrest via inhibition of cyclin dependent kinase activities in many breast cancer cell lines [30]. Reprimo, a highly gylcosylated protein and p53 regulated G2/M progression regulator, is also a candidate gene that is involved in G2/M cell cycle arrest. Ectopic expression of p53 protein induced its mRNA expression and subsequently induced G2/M cell cycle arrest. In arrested cells, Reprimo is able to inhibit the activity of cdc2 and nuclear translocation of cyclin B1. Therefore, Reprimo is considered to be another p53-induced cell cycle regulator [31]. Recently, Reprimo has been identified as a potential biomarker for early detection of gastric cancer [32]. Besides, loss of Reprimo expression due to aberrant methylation was identified in various human malignancies including gastric, gallbladder, colorectal, breast, pancreatic and lung cancers, leukemias, lymphomas and esophageal adenocarcinomas [33,34]. However, little is known about this gene in EBV-associated cancers.
Previous studies showed that LMP-1 could activate cell cycle regulator through activation of NFκB and AP-1 pathway and thus induced obstruction in cell cycle [35,36]. In the present study, we aim to express the LMPs and identify the target genes that induce G2/M arrest in LMP-1 expressing cell as well as the interaction of LMPs co-expression. Since 14-3-3σ and Reprimo are well-known G2/M regulator that have been associated with various human cancers, we hypothesized that these two genes are involved in LMP-1-induced G2/M arrest. Therefore, we examined the expression of these two genes in LMPs expressing cells.
2 Materials and methods
2.1 Cell lines and cell cultures
The B95.8 cell line was generous gift from Dr. Sam Choon Kook; the HEK293T line obtained from ATCC®, USA (CRL-11268™). The B95.8 cell line was established by infection of EBV to marmoset monkey (Saguinus Oedipus) blood leukocytes. The B95.8 cell line was used as a source for LMP-1, LMP-2A and LMP-2B genes. B95.8 was maintained in RPMI-1640 supplemented with 10% v/v FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin (GIBCO/BRL, Grand Island, NY). HEK293T cells which are widely used in molecular biology research for many years due to its viability and high transfection efficiency was maintained in DMEM supplemented with 10% v/v FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin. All cells were maintained in an incubator (NuAire Inc., USA) at 37 °C in 5% CO2 and sub-cultured in 1:10 ratio in the same condition unless for specific application.
2.2 Construction of expression plasmids
RNA was extracted from cells that have been stabilized in RNAlater® using the RNeasy Protect Kit® (Qiagen, Germany) according to manufacturer's protocol. Total RNA from B95.8 cells was subjected to reverse transcription (RT) to generate cDNA of LMP-1, LMP-2A and LMP-2B. Reverse transcription was performed using SuperScript™ III First-Strand Synthesis System (Invitrogen, USA) following the manufacturer's protocol. The LMP-1, LMP-2A and LMP-2B amplified products were cloned into pcDNA™ 3.1 Directional TOPO® vector (Invitrogen, USA). For tagging of LMP-2B protein with V5 epitope and polyhistidine region, the termination codon was removed from the PCR primer. The pcDNA3.1 vector with correct insertion was confirmed by sequencing. Then, the sequences were compared and aligned with B95.8 sequence from NCBI (Gene Bank Acc. No.: V01555).
2.3 Cell transfection
LMP-1, LMP-2A and LMP-2B expressing cells were established by transfecting the cells with pcDNA3.1-LMP-1, pcDNA3.1-LMP-2A and pcDNA3.1-LMP-2B-V5 constructs. The fluorescence protein expression vector, pcDNA3.1-CFP, was used to monitor the transfection efficiencies. Transfection of the epithelial cells was performed by Fugene® HD Transfection Reagent following manufacturer's protocol. Briefly, 1 × 106 cells were seeded in T-25 cell culture flask in DMEM-10 with no antibiotic and incubated for 24 h at 37 °C with 5% CO2. For transfection, 6 μg of plasmid (3 μg of pcDNA3.1-empty vector + 3 μg of pcDNA3.1-LMP-1 or pcDNA3.1-LMP-2A or pcDNA3.1-LMP-2B for the respective LMPs expression; 3 μg of pcDNA3.1-LMP-1 + 3 μg of pcDNA3.1-LMP-2A or pcDNA3.1-LMP-2B-V5 for co-expression of LMPs) was added into 600 μl of opti-MEM® medium. Subsequently, the diluted plasmid was added with 30 μl of Fugene® HD Transfection Reagent (Roche, Switzerland) in a ratio of 1:5 and mixed well. The mixture was incubated for 15 min at room temperature for transfection-complex formation. Next, the transfection-complex was added into the seeded cells in drop-wise manner. The cells were incubated for 48 h at 37 °C with 5% CO2. After 48 h incubation, the cells were harvested for other assays.
2.4 Analysis of gene expression in LMP-1, LMP-2A and LMP-2B expressing cells
2.4.1 Real-time quantitative PCR
Real-time quantitative PCR was performed in a volume of 15 μl reaction which contained 100 ng of cDNA template, 1 × SYBR Green PCR Master mix and 0.2 mM forward and reverse gene specific primers using ABI 7000 Real-time PCR machine (Applied Biosystem, USA). Each PCR reaction was optimized to eliminate the presence of primer dimer. For each pair of genes specific primers, three independent RT-PCRs were performed using three different sets of RNA samples as template. The PCR conditions were as follows: 95 °C for 10 min followed by 40 cycles of 94 °C for 15 s and 60 °C for 1 min. The fluorescent product was determined at amplification step of each cycle. After amplification, melting curve was carried out by heating the PCR product to 94 °C for 5 min and cooling at 0.5 °C/10 s to 55 °C. The fluorescence data was collected in 0.5 °C interval.
The relative fold change of the gene of interest was determined by comparing its expression level to GAPDH or β-actin internal control as follows: the relative fold change = 2−ΔΔCq, where ΔΔCq = [[CqTarget (sample) – CqGAPDH (sample)] – [CqTarget (Empty vector) – CqGAPDH (empty vector)] + [CqTarget (sample) – Cqβ-actin (sample)] – [CqTarget (Empty vector) – Cqβ-actin (empty vector)]]/2.
Primers such as GAPDH (Forward-5’ CCA CCC ATG GCA AAT TCC 3’ and Reverse-5’ CAG CAT CGC CCC ACT TG 3’), β-actin (Forward-5’ AAA AGC CAC CCC ACT TCT CTC T 3’ and Reverse-5’ AAT GCT ATC ACC TCC CCT GTG T 3’), 14-3-3σ (Forward-5’ TGT CCA GTA TTG AGC AGA AAA GCA 3’ and Reverse-5’ CAC GCC CTG GAG CTC AGT 3’) and Reprimo (Forward-5’ CTG GCC CTG GGA CAA AGA C 3’ and Reverse-5’ TCA AAA CGG TGT CAC GGA TGT 3’) were designed with Primer express 3.0.
2.4.2 SDS-PAGE and Western Blotting
The cell lysates were prepared using Radio-Immunoprecipitation Assay (RIPA) buffer: 50 mM Tris-Cl (Sigma, USA), 150 mM NaCl (Merck, Germany), 0.5% w/v Na-deoxycholate (Sigma, USA), 0.1% w/v Sodium dodecyl sulphate (SDS) (Amresco, Ohio), 1% v/v NP-40 (Sigma, USA) with supplement of protease inhibitor cocktail (Calbiochem, La Jolla, CA). Cell lysate was quantified with Bradford's method with improved dye reagent from Bio-Rad. The extracted proteins were mixed with 6 × sample treatment buffer and heat denatured at 94 °C for 5 min (at 70 °C for 5 min for LMP-2A and LMP-2B). Fifty micrograms of the protein was loaded on a 12% SDS polyacrylamide gel for separation. After separating proteins using SDS-PAGE, the proteins were transferred electrophoretically to a PVDF membrane using Trans-Blot® SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). The primary antibodies used were CS1-4 LMP-1 (1:1000) (Dako, Denmark), B14-7 LMP-2A (1:1000) (Serotec, UK), Anti-V5 (1:1000) (Invitrogen, USA) for LMP-2B, Reprimo (1:1000) (Genetex, USA) and Alpha-Tubulin (1:1000) and 14-3-3 Sigma (1:1000) (Abnova, Taiwan). The secondary antibodies used were HRP-anti-mouse/rat (1:2000) (Dako, Denmark), AP-anti-mouse (1:5000) (Sigma, USA) and HRP-anti-rabbit (1:1000) and anti-goat (1:1000) (Abnova, Taiwan). The protein signal was detected with BCIP/NBT phosphate substrate system for AP-conjugated secondary antibody. For HRP-conjugated secondary antibody, protein signal was detected by Western Lightning® Plus–ECL detection system according to manufacturer's protocol.
2.5 Flow cytometry
Forty-eight hours after transfection, the cells were harvested prior to propidium iodide staining. The cells were trypsinized and washed twice with PBS, then collected in 15 ml round bottom tube (Becto Dickinson, San Jose, CA). Next, the cells were fixed in ice-cold absolute ethanol at 4 °C for 1 h and then collected by centrifugation. The pellet was washed once with PBS and incubated for 5 min on ice in 5 ml of PBS with 0.25% Triton-X 100. The cells were then collected and incubated in PBS containing 10 μg/ml propidium iodide and 10 μg/ml RNaseA at 4 °C for 20 min. The cells were kept in 4 °C prior to FACS analysis using FACS Calibur flow cytometer (Becto Dickinson San Jose, CA). The histograms from FACS were further analyzed with winmbi and Cylchred cell cycle analysis software.
2.6 Data analysis
Graphpad PRISM 5.0 was used to evaluate the collected data. Results were evaluated by Anova. All statistical tests with P-values < 0.05 were considered statistically significant.
3 Results
3.1 Expression of LMP-1, LMP-2A, LMP-2B in HEK293T cell line
By RT-PCR, the expressions of LMP-1, LMP-2A and LMP-2B mRNA were confirmed in HEK293T cells (Fig. 1A). The cDNAs from transfected cells were subjected to RT-PCR using gene specific primers and the products were analyzed via agarose gel eletrophoresis. As shown in Fig. 1, all the transfected cells expressed LMPs at a detectable level except cells transfected with pcDNA3.1-empty vector.
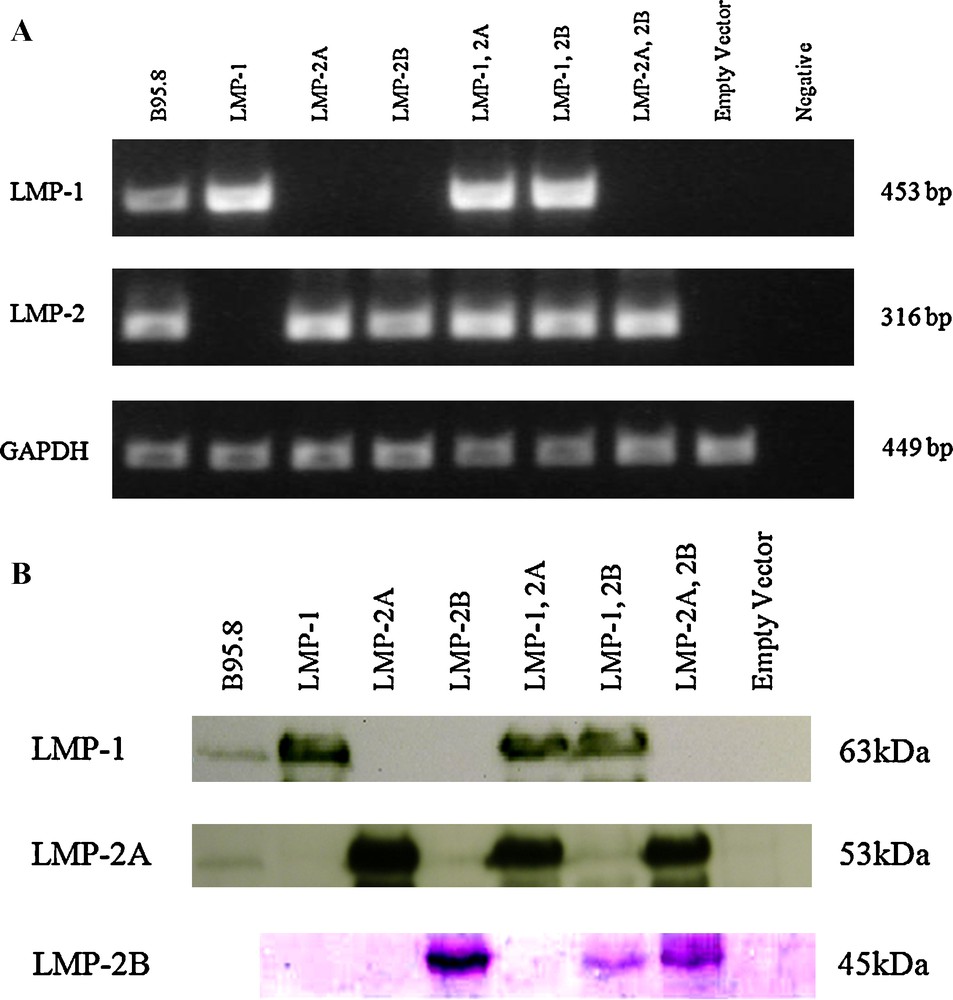
Expression of LMP-1 and LMP-2 in HEK293T. (A) Both of LMP-2A and LMP-2B share a common sequence therefore only 1 pair of primer was used to verify the expression of LMP-2. Same amount of cDNAs were subjected to semi-quantitative PCR and the amplified products were analyzed in 1% agarose gel electrophoresis. The cDNA from B95.8 was used as a positive control for LMPs gene expression. GAPDH was used as internal control for cDNA abundance normalization. (B) Fifty μg of total protein lysates were separated in 10% SDS-PAGE and transferred to PVDF membrane for blotting with specific antibody. B95.8 cell lysate was used as a positive control for LMP-1 and LMP-2A proteins whereas LMP-2B was detected with tagged epitope (V5).
The expression of LMPs proteins in the transfected cells was detected with western blot assay (Fig. 1B) 48 h after transfection. Western blot analysis revealed that LMPs with the molecular weights of 63 kDa, 53 kDa and 45 kDa were expressed in cells transfected with pcDNA3.1-LMP-1, pcDNA3.1-LMP-2A and pcDNA3.1-LMP-2B-V5 constructs respectively. These observations indicated that the expression vectors were successfully transfected into cells and expressed LMPs at detectable levels.
3.2 Reduction in cell density in all LMP-1 expressing cells
Forty-eight hours post-transfection, cells were examined microscopically for effects of LMPs expression on cell viability. LMP-1 is toxic to cells and causes cytostasis when expressed in high levels [20,25]. We observed reduction in cell density in all LMP-1 expressing cells (Fig. 2) 48 h post-transfection. There were less or no obvious phenotypical changes observed in these cells compared to their control cells. The control cells (pcDNA3.1-empty vector) showed a typical appearance of epithelial cells with flattened cell morphology and exhibited clear cell-to-cell contact and adhesion.
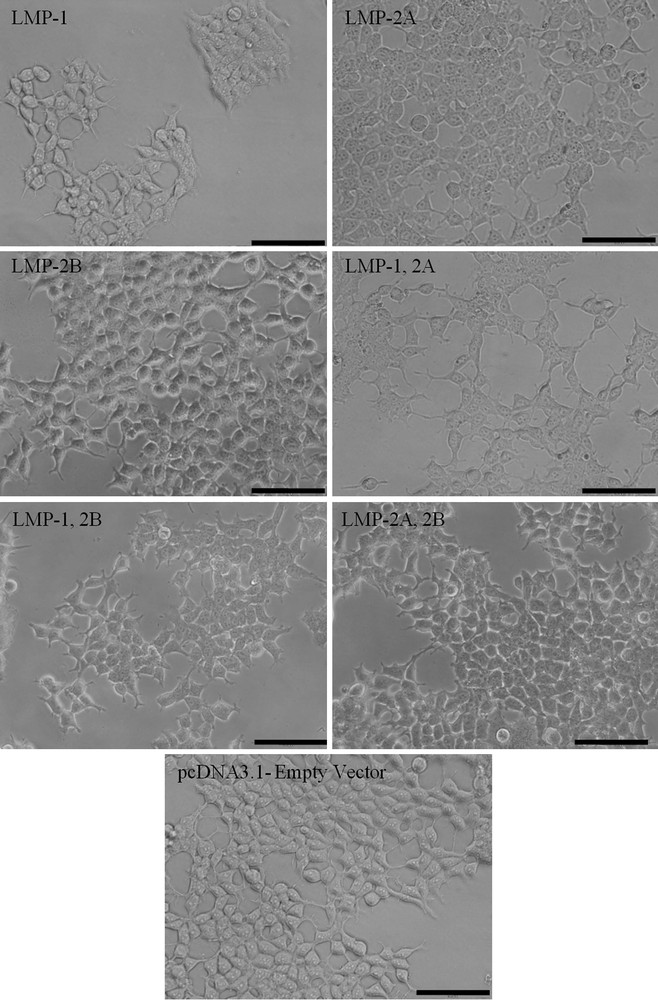
Effects of LMPs expression on cell viability and morphology. The LMP-1 expressing cells contain lesser cells compared to the cells transfected with empty vector (Scale bar, 100 μm).
3.3 Upregulation of 14-3-3σ and Reprimo in LMP-1 expressing cells
The upregulation of 14-3-3σ and Reprimo was further analyzed with real-time PCR assay. GAPDH and β-actin were used as endogenous controls. Values for Relative Quantitative (RQ) were determined from the Cq of each reaction. In order to determine specific amplification, melting curves of the PCR products were studied and only single peak for all amplifications were observed. Upregulation of these genes were confirmed in all the LMP-1 expressing cells (Fig. 3). The gene expression of 14-3-3σ was upregulated in LMP-1, LMP-1, 2A and LMP-1, 2B expressing cells by 3.49 fold (P < 0.001), 2.78 fold (P < 0.001) and 2.39 fold (P < 0.01), respectively when compared to the empty vector. Reprimo was upregulated by 2.08 fold (P < 0.001), 2.27 fold (P < 0.001) and 1.76 fold (P < 0.01), respectively when compared to the empty vector.
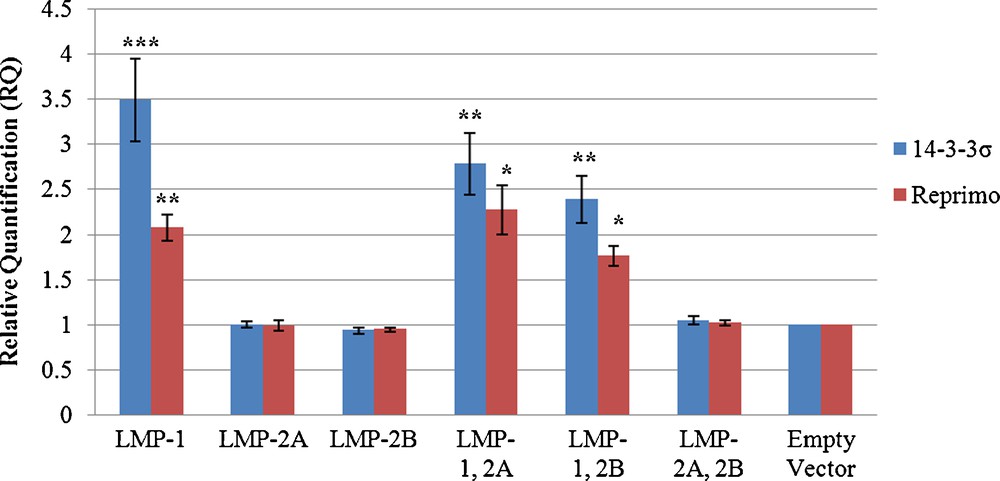
The mRNA expression of 14-3-3σ and Reprimo in LMPs expressing cells. Relative gene expression levels for all clones were determined using real-time quantitative analysis. Data shown are the mean of three independent experiments with standard error as error bars. *Indicates where value is significant in comparison to negative control (pcDNA3.1-empty vector) (Student's t-test, ***P < 0.001, **P < 0.01 and *P < 0.05).
3.4 14-3-3σ and Reprimo proteins were upregulated in all the LMP-1 expressing cells
In this study, the protein expression levels of 14-3-3σ and Reprimo were further examined with specific antibodies via western blotting analysis. As shown in Fig. 4, protein levels of 14-3-3σ and Reprimo were upregulated in all the LMP-1 expressing cells.
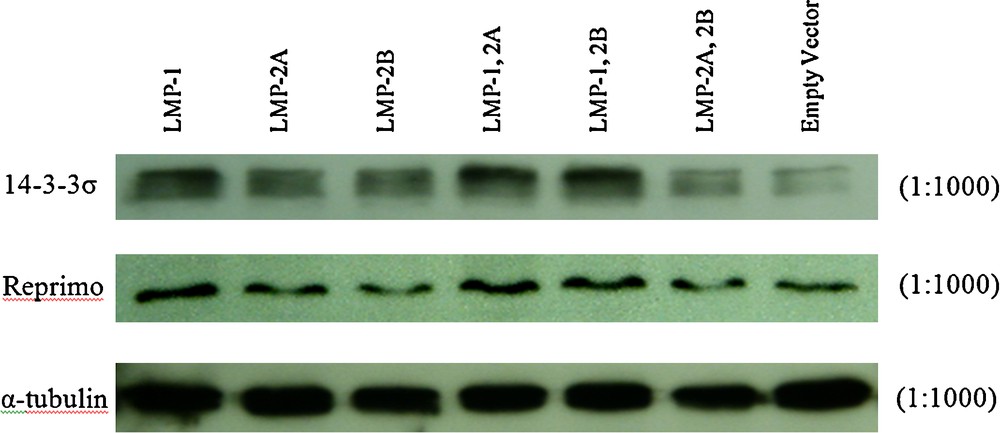
Western blotting analysis of 14-3-3σ and Reprimo in various clones. Cells transfected with empty vector were used as a negative control. Fifty μg of total protein lysates were separated in 12% SDS-PAGE and transferred to PVDF membrane for blotting with specific antibodies. Protein levels for 14-3-3σ and Reprimo in the various clones were shown and α- tubulin protein level was detected as loading control.
3.5 Overexpression of LMP-1 arrests cells in G2/M phase
In order to understand the effects of LMPs expression on cell cycle progression, transfected cells were subjected to flow cytometry analysis based on DNA content in nuclei stained by PI. Forty-eight hours post-transfection, cell cycle analysis revealed an increase in G2/M phase in all the LMP-1 expressing cells compared to the control cells (Fig. 5). Our data has shown that about 25.13 ± 3.93% (P < 0.05) of the cells expressing only LMP-1 accumulated in G2/M phase 48 h after transfection, in contrast to only about 16.73 ± 0.26% of negative control cells (pcDNA3.1-empty vector) in the same phase. When LMP-1 was co-expressed with LMP-2A or LMP-2B, the proportion of cells accumulated in G2/M phase were 25.93 ± 1.07% (P < 0.01) and 24.97 ± 3.88% (P < 0.05), respectively (Fig. 5B and Table 1). The results revealed that the overexpression of LMP-1 capable to arrest the cells in G2/M phase. Details of cell cycle distributions for transfected cells and HEK293T cells are summarized in Table 1.
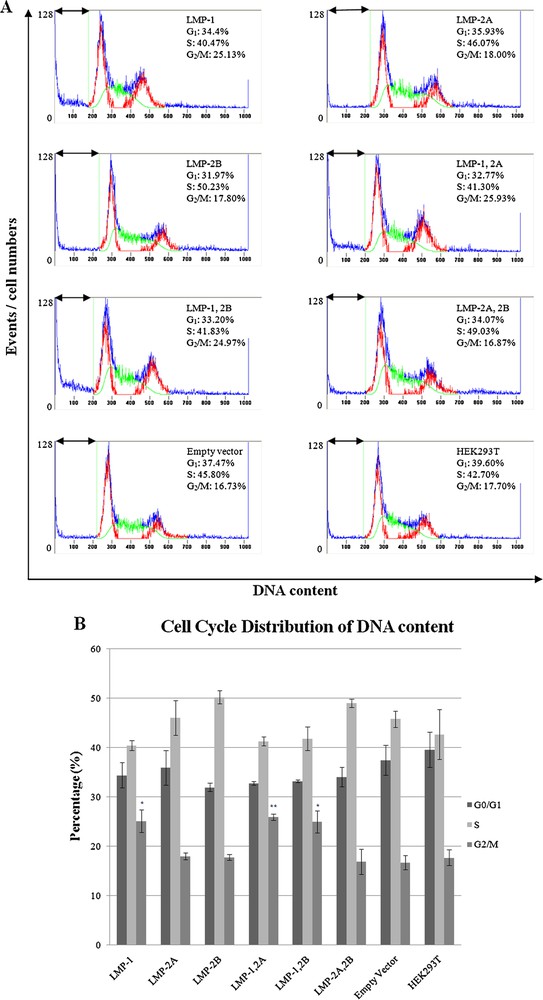
Flow cytometry analysis of HEK293T cells. (A) Histograms represent cell cycle distributions of HEK293T cells transfected with pcDNA3.1 and non-transfected parental cells. DNA profiles were analyzed using Cylchred software. G1 and G2/M phases are shown in red, and S phase is shown in green. The arrow indicates the apoptotic-sub G1 fraction of the cells. (B) Cell cycle phase distribution was analysed. Transfected cells were compared to cells transfected with pcDNA3.1-empty vector (mock). Data shown are the mean of three independent experiments with standard error as error bars. *Indicates where value is significant in comparison to negative control (pcDNA3.1-empty vector) (ANOVA, **P < 0.01; *P < 0.05).
Cell Cycle distribution of transfected cells and HEK293T cells.
| Clones | G0/G1 (%) | SD (%) | S (%) | SD (%) | G2/M (%) | SD (%) | Total |
| LMP-1 | 34.40 | 4.43 | 40.47 | 1.82 | 25.13* | 3.93 | 100.00 |
| LMP-2A | 35.93 | 6.09 | 46.07 | 6.03 | 18.00 | 1.23 | 100.00 |
| LMP-2B | 31.97 | 1.50 | 50.23 | 2.37 | 17.80 | 0.95 | 100.00 |
| LMP-1, 2A | 32.77 | 0.67 | 41.30 | 1.61 | 25.93** | 1.07 | 100.00 |
| LMP-1, 2B | 33.20 | 0.46 | 41.83 | 4.11 | 24.97* | 3.88 | 100.00 |
| LMP-2A, 2B | 34.07 | 3.36 | 49.03 | 1.52 | 16.87 | 4.47 | 100.00 |
| Empty Vector | 37.47 | 5.48 | 45.80 | 5.51 | 16.73 | 0.26 | 100.00 |
| HEK293T | 39.60 | 6.19 | 42.70 | 8.70 | 17.70 | 2.74 | 100.00 |
4 Discussion
LMP-1 is a well-known oncogene that commonly causes tumorigenesis by activating multiple signaling pathways in ligand independent manner. It can trigger wide range of signaling pathway which is involved in cell growth, apoptosis and differentiation. For example, LMP-1 activates PI3K/Akt with its CTAR1 region which is important to promote cell survival [37]. Moreover, it can activate MAPK, plays an important role in coordinating cellular activities from gene expression, mitosis, and metabolism to survival, apoptosis and differentiation [38]. On the other hand, LMP-1 is able to inhibit cell proliferation or induce cytostatic effect when expressed efficiently [20,23,25,39]. LMP-1 was expressed in one to four times higher than EBV-positive B-cells and induced cytostasis in 293 cells for at least 6 days [39]. Floettmann and co-workers also showed that LMP-1 induced cytostatic effect in BJAB, DG75 and Akata cells but some of the cells resumed proliferation after 4 days [25]. The recovered cells may be part of the cells with lower expression level of LMP-1. Hence, LMP-1 may have growth advantage in those cells with lower expression levels. In our present study, LMP-1 expressing HEK 293T cells exhibited cytostatic effect when expressed in higher level compared to its expression level in B95.8 cells. This was evident from the observation of a reduction in cell density in the LMP-1 expressing cells. Reduction in cell density may be due the apoptosis and/or cell cycle arrest. Part of the cells may have undergone apoptosis, since we observed an increase in sub G1 population. Moreover, in agreement with Floettmann et al. [25] and Deng et al. [24], our flow cytometry analysis revealed that LMP-1 arrested cells at the G2/M phase of the cell cycle.
There are possible consequences of this LMP-1 induced cytostasis effect. Upon infection of primary B-cells, EBV DNA is increased and this amplification is thought to be vital in maintaining an optimal level of viral expression. Eventually, the number of DNA copies stabilizes. The stabilization may be caused by LMP-1's role in growth inhibition whereby a higher LMP-1 expression level restricts cell proliferation [39]. In Burkitt's lymphoma, one of the fastest proliferating human tumors, the expression of LMP-1 is lost [40]. The rapid proliferation of this tumor may be contributed by the loss of LMP-1's growth inhibitory properties. Lastly, the susceptibility of EBV infected cells to exterior cytotoxic signals may be limited by the system which LMP-1 employs to induce cytostasis [39]. LMP-1 is a well-known oncogene but its cytostatic effect may be an important feature for cancer cells to resist chemotherapy and radiotherapy. Moreover, arrested cells may be protected from apoptosis and therefore be resistant to the cytotoxic agent [41] as well as radiotherapy [42].
In our study, we identified another two downstream targets of p53 which are 14-3-3σ and Reprimo that may be involved in G2/M arrest in LMP-1 expressing cells. p53 protein is disabled in 293T cells by E1B and T large antigen; therefore we suggested that 14-3-3σ and Reprimo maybe upregulated via p53-independent pathway. According to Zhang et al. [43], 14-3-3σ is found to be stimulated by PI3K signaling pathway when in response to IGF-I. They have shown that IGF-I induced expression of 14-3-3σ consistently even when p53 expression was knockdown with siRNA in MCF-7 cells. Therefore, we think that LMP-1 may probably bypass p53 to induce 14-3-3σ expression in HEK293T cells since LMP-1 is involved in many signaling pathways. However, the exact mechanism still needs further investigation. However, Reprimo expression in p53-independent manner has not been well studied; hence this occurrence may need additional studies.
On the other hand, since LMPs are commonly co-expressed and co-localized to the same compartment [15–17], they may probably interact with each other in tumorigenesis. However, in our present observation, LMP-2A and 2B were found to have no significant changes on LMP-1 induced 14-3-3σ and Reprimo expression and G2/M arrest. This observation still needs further investigation to unveil the mechanism of LMPs interaction.
In conclusion, we have shown that LMP-1 may induce cell cycle arrest in G2/M phase through upregulation of 14-3-3σ and Reprimo in p53-independent manner. Moreover, LMP-2 may not interfere with the function of LMP-1 in inducing cell cycle arrest. According to our study, LMP-1-induced 14-3-3σ and Reprimo may play some roles in EBV-associated human malignancies. Further studies will be required to determine the precise machinery by which LMP-1 induces cell cycle arrest for better understanding of this distinct mechanism.
Disclosure of interest
The authors have not supplied their declaration of conflict of interest.
Acknowledgements
This research work was funded by eScience fund (02-01-03-SF0007) from MOSTI, PPP Grant (PS283/2009B & PS271/2010B) and High Impact Research Grant (UM.C/625/1/HIR/MOHE/CHAN-02 (H-50001-A000023) from University of Malaya.


