1 Introduction
The longface emperor, originally described by A. Valenciennes [1] as the “léthrinus olive” from a specimen collected in the Sunda Strait by J.R.C. Quoy and P. Gaimard is the largest and the longest-snouted lethrinid [2,3]. This carnivorous fish, whose adult size commonly reaches 70 cm, dwells on the sandy shores, reef slopes and lagoons of the tropical Indo-West Pacific [2–6]. It is a species of prime commercial importance throughout the region [2]. Among the many other Lethrinus spp. species described by A. Valenciennes [1], three, namely L. rostratus, from the East Indies (“Batavia”), L. waigiensis from Waigeo Island, West Papua, and L. xanthopterus from Ulea, Caroline Islands, represent the same species and have all been synonymised with L. olivaceus [2]. The existing type material for these species actually consists of individuals of small size that have been poorly preserved, except the holotype of L. olivaceus ([2]; Fig. 1). Given the circumstances, meaningful morphological differences between these type specimens, if any, arguably may have gone unnoticed. No type specimen is known for L. xanthopterus and A. Valenciennes’ short description of this species only refers to “les riches collections de dessins que MM. de Mertens et Ketlitz ont bien voulu nous communiquer à leur retour du dernier voyage des Russes autour du monde” [1]. R.L. Playfair and A.C.L.G. Günther later described L. longirostris from Zanzibar [7] which was eventually considered as yet another junior synonym of L. olivaceus [2,8].
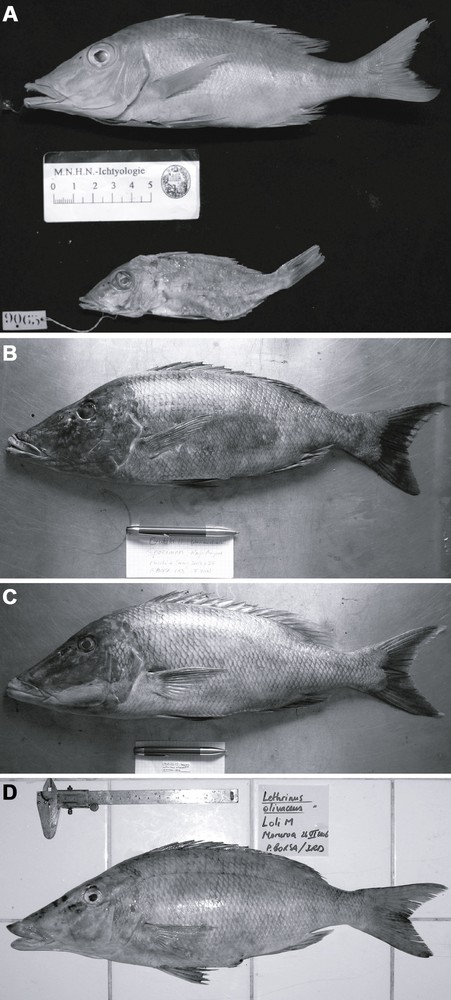
Lethrinus olivaceus type material and voucher specimens. A. Above: holotype of L. olivaceus Valenciennes 1830 (specimen MNHN 0000-5741), from Antjer, Sunda Strait, Indonesia [1]; standard length (SL) 175 mm. Below: holotype of L. waigiensis Valenciennes 1830 (specimen MNHN 0000-9065), from Waigeo, West Papua [1]; SL 107 mm (PB). B. Voucher specimen Gutila 11 (LIPI-P2O, Jakarta, unregistered) from Raja Ampat, West Papua, November 2007; SL 442 mm; of the “Pacific Ocean” form (or Lethrinus sp. B) according to nucleotide sequence at the cytochrome b locus (GenBank KC596047) (PB). C. Voucher specimen Gutila 12 (skull preserved at IRD-UR 227, Montpellier) from Raja Ampat, West Papua, November 2007; SL 539 mm; of the “Coral Triangle” form (or Lethrinus sp. A) according to nucleotide sequence at the cytochrome b locus (GenBank KC596051) (PB). D. Specimen MNHN 2009-0016 from Moruroa, French Polynesia, June 2006; SL 382 mm; of the “Pacific Ocean” form (or Lethrinus sp. B) according to nucleotide sequence at the cytochrome b locus (GenBank KC596057) (PB).
A phylogenetic tree based on nucleotide sequence variation of the mitochondrial control region in several Lethrinus species illustrated a deep split between L. olivaceus haplotypes from Bali and New Caledonia [9], suggesting the possible occurrence of a cryptic species. In this study, we further investigated morphological variation in L. olivaceus. Mitochondrial DNA haplotype (cytochrome b gene) sequences for samples from these and from additional locations throughout the Indo-West Pacific were also scored. The combined evidence from morphology and phylogeography allowed us to address taxonomic issues in this species.
2 Materials and methods
Fifty-four Lethrinus olivaceus individuals were sampled in 2006–2011 from ten localities in the Indo-West Pacific (Fig. 2) as part as an ongoing program on the multiple-species phylogeography of the Lethrinidae. Identification to species was made on the basis of the distinctive morphological and meristic features given by K.E.C. and G.R. Allen [2], which include the ratio of head length to body depth and the count of scale rows between the lateral line and the basis of the first dorsal fin. Voucher specimens (Fig. 1, Appendix 1) included one individual from Sorong (West Papua) shipped frozen to LIPI-Oseanografi in Ancol (Indonesia), one specimen from Moruroa (French Polynesia) shipped frozen to IRD UR 227 in Nouméa (New Caledonia) and subsequently fixed in formalin and deposited at Museum national d’histoire naturelle, Paris (France) under collection no. MNHN 2009-0016, and the skulls and jawbones of 36 individuals from the Maldives, Bali, West Papua, New Caledonia, and Moruroa, now preserved at IRD-UR 227 in Montpellier (France). Two L. microdon individuals were also sampled from the Baa atoll (Maldives) and the Kedonganan fish market (Bali), respectively.
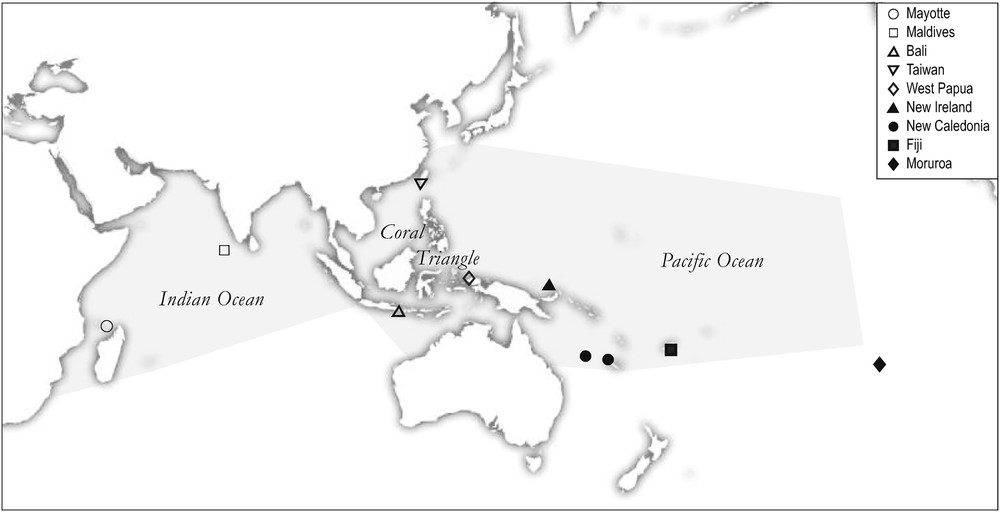
Longface emperor, Lethrinus olivaceus. Sampling locations of individuals selected for morphometric analysis and/or genetics. Mayotte: Bancs Geyser and Zélée, Mozambique Channel, April 2011 (N = 2); Maldives: Baa atoll, June 2008 (N = 4); Bali: Kedonganan fish market, February 2007 (N = 5); Taiwan: Kenting, May-June 2011 (N = 4); West Papua: Sorong, November 2007 (N = 5); New Ireland: Kavieng, August 2006 (N = 1); New Caledonia: Northern lagoon, April 2007 (N = 22) and Chesterfield Islands, Coral Sea, August 2011 (N = 1); Fiji: Tai Levu, September 2008 (N = 1); Moruroa: Moruroa lagoon, June 2006 (N = 8). Shaded: distribution of the species, as inferred from the point map provided by FishBase [10] (accessed 02 November 2012).
The samples analyzed for morphometric variation consisted of heads or carcasses of L. olivaceus purchased from the Male (Maldives) (N = 1), Kedonganan (N = 4), Sorong (N = 1), and Nouméa (N = 22) fish markets in 2007–2008. To prepare the neurocrania and the jawbones, the specimens were boiled, and the boiled skin and flesh were then separated from the bone using forks, fingers, twizzles, toothpicks, toothbrushes and a wash bottle. The specimens sampled in the Moruroa lagoon, June 2006 (N = 8) were shipped frozen to Noumea and the heads were spared for similar processing. Neurocrania of individuals from Bali ranged in condylo-basal length from 81.2 mm to 111.9 mm; those from New Caledonia, from 80.9 mm to 102.2 mm; and those from Moruroa, from 86.6 mm to 126.3 mm. The neurocranium of the single specimen examined from the Maldives was 130.8 mm long; that of the single specimen examined from West Papua was 129.8 mm long. Nine morphometric variables (Fig. 3A) were measured on the neurocranium and jawbones. To normalize the variables and standardize them for size, prior to analysis, each was log-transformed and divided by the log-transformed condylo-basal length [11,12]. We used principal component analysis (PCA) [13] to determine whether longface emperor populations were craniometrically distinct. Principal component analysis was run using the ADE-4 software [14].
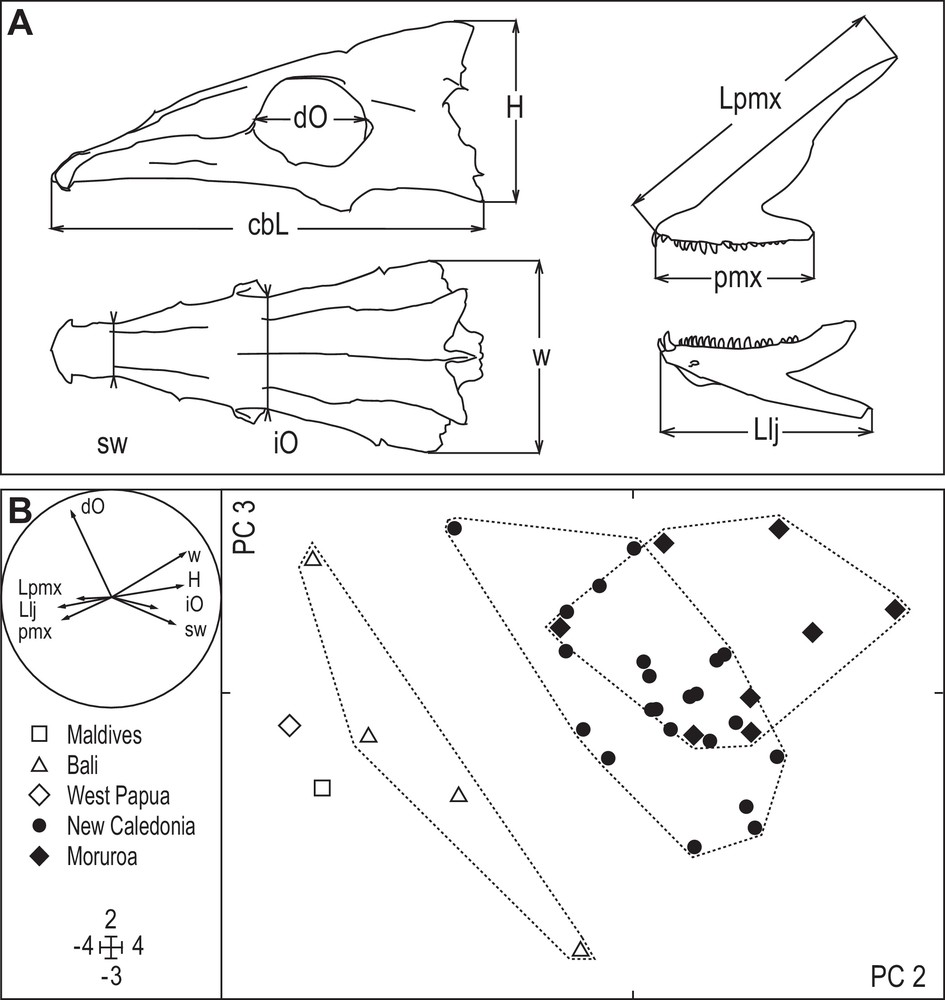
Longface emperor, Lethrinus olivaceus. Morphometric analysis of head skeleton. A. Measurements on neurocranium, left pre-maxilla (upper jaw) and left lower jaw, made to the nearest 0.1 mm using a Vernier calliper: cbL, condylo-basal length; dO, orbit diameter; H, neurocranium height; iO, interorbital width; w, neurocranium width; sw, width of snout where narrowest; Lpmx, length of premaxilla; pmx, length of toothed part of premaxilla; Llj, length of lower jaw. To normalize the variables and standardize them for size, prior to analysis each was log-transformed and divided by the log-transformed condylo-basal length [11,12]. B. Principal component analysis (output from ADE-4 [14]): scores for principal components (PC) 2 (horizontal: 23.6% of total variance) and 3 (vertical: 10.6% of total variance) of individuals from the Indian Ocean (Maldives: N = 1), the Coral Triangle (Bali: N = 4; West Papua: N = 1) and the Pacific Ocean (New Caledonia: N = 22; Moruroa: N = 8), characterized by the above-defined morphometric variables. Inset: correlation circle [14]; legend to symbols; graduation on axes.
The choice of the neurocranium (instead of, e.g., the whole fish) to study geographic variation in morphology in L. olivaceus was dictated by site accessibility and size of specimens. In some of the places where they were sampled (Maldives and New Caledonia), emperors were filleted and the carcasses were sold separately, hampering the provision of entire specimens. Specimens of this species are usually of such large size that preserving them whole and shipping them poses a challenge when operating from remote field sites. Neurocrania, however, have the advantage that once cleaned and dried, they do not necessitate specific preservation conditions nor much storing space.
Fin clips of a number of samples (N = 28; Appendix 1) were preserved dry or in 95% ethanol and an aliquot of these was shipped to the Marine Phylogenomics Laboratory, Taipei, for DNA extraction and sequencing analysis. DNA was extracted from subsamples of the fin clips using the Qiagen DNAeasy extraction kit (Qiagen, Valencia CA, USA) according to the manufacturer's instructions. The concentration of extracted DNA was measured by spectrophotometry using an Eppendorf spectrophotometer (Eppendorf, Hamburg, Germany). Conditions for amplification by polymerase chain reaction (PCR) were as follows: 0.5 U GoTaq® Flexi DNA polymerase (Promega, Madison, U.S.A.), 1× reaction buffer, 2 mM MgCl2, 200 μM each dNTP, 0.2 μM each primer, and 20–50 ng genomic DNA in a 25-μl final reaction volume. Primers MT14673F (5′-TAACCAGGACTAATGGCTTGAAA-3′) and Let15856R (5′-GAGCTACTAGTGCATNGTCAKCC-3′) designed by W.-J.C. (this study) were used for amplifying the entire cytochrome b gene. Thermocycling conditions for PCR were: initial denaturing step at 95 °C for 4 min followed by 35 cycles of 95 °C (for 40 s), annealing at 55 °C (for 40 s), and 72 °C (for 1.5 min), and a final extension step of 72 °C (for 7 min) before thawing at 4 °C. The PCR product was purified using AMPure magnetic beads (Agencourt Bioscience Corp., Beverly, MA, USA) and resuspended in 30 μL of sterile water. Nucleotide sequences of the PCR-amplified DNA fragments were then determined using the sequencing facilities of the National Taiwan University and Genomics Inc. (Taipei, Taiwan) using ABI 3730xl analyzers (Applied Biosystems, Foster City CA, USA). Nucleotide sequences were edited and managed using the CodonCode Aligner v. 3.0.7 (CodonCode Corporation, Centerville MA) and Se-Al v. 2.0a11 [15] together with sequences AF381265 (L. microdon), AF381252 (L. olivaceus), AF381253 (L. reticulatus), and AF24075 and AF381263 (L. rubioperculatus) retrieved from GenBank (http://www.ncbi.nlm.nih.gov). The final matrix of aligned sequences used for phylogenetic analysis, representing a total of 33 haplotypes, was 1107 base pairs in length. L. microdon, L. reticulatus and L. rubrioperculatus were selected as outgroups because of their close evolutionary affinity to L. olivaceus [16]. Phylogenetic analyses were based on a partitioned maximum likelihood (ML) method as implemented in RAxML 7.0.4 [17]. A mixed model analysis was used that allows the independent estimation of individual models of nucleotide substitution for each partition (by codon position). A GTR+G model for each partition was used and an Optimal ML tree search was made with 100 separate runs using the default algorithm of the program from a random starting tree for each run. The final tree was selected among suboptimal trees in each run by comparing likelihood scores under the GTR+G model. To evaluate the robustness of the internal branches of the ML tree, 1,000 bootstrap replications were run for the data set under the GTR+G model. Ancillary descriptive statistics were computed using the Free Statistic Software (Calculator) software [18] and the Mega5 software [19].
3 Results
Values for cranial morphometric variables in 36 L. olivaceus specimens are presented in Appendix 2. Principal component (PC) 1 (not shown) from PCA on cranial morphometric variables was correlated with condylo-basal length (Spearman's rank correlation [18,20]: ρ = 0.838; P < 0.001). This suggests a strong and expected effect of growth allometry. The pattern defined by PC 2 and PC 3 (Fig. 3B) segregated the individuals by geographic origin. All individuals from New Caledonia and Moruroa (Pacific Ocean) were grouped into a single cluster, albeit with some statistical differences between the two populations. Individuals sampled in Bali were clustered separately (Fig. 3B). The single specimen from West Papua and the single specimen from the Maldives were positioned close to the Bali cluster along PC 2 (Fig. 3B).
Differences in neurocranial morphology are likely to translate into differences in head shape. The few pictures available (some of which are presented in Fig. 1) suggest that Lethrinus sp. B has a more slender snout than Lethrinus sp. A.
Cytochrome b gene sequences from 27 L. olivaceus individuals sampled across the species range allowed the reconstruction of the phylogeny of mitochondrial lineages distribution-wide (Fig. 4). Two major clades were observed: one clade comprised mitochondrial haplotypes sampled from the Pacific Ocean, including two locations from the eastern Coral Triangle (Taiwan and West Papua); the other clade consisted exclusively of haplotypes sampled in the Indian Ocean (forming an exclusive sub-clade) and in the Coral Triangle (forming another sub-clade). The specimen from West Papua that was used for the morphometric analysis (Gutila 12: Appendices 1–3) was found to belong to the “Coral Triangle” sub-clade. The net nucleotide distance (Kimura-2 parameter distance [19]) between the “Pacific Ocean” clade and the (“Indian Ocean” + “Coral Triangle”’) clade of L. olivaceus was 0.095. The net distance between the “Indian-Ocean’ and “Coral-Triangle” subclades was 0.017. Nucleotide sites diagnostic between the “Pacific Ocean” clade vs. the (“Indian Ocean” + “Coral Triangle”) clade are highlighted in Fig. 5.
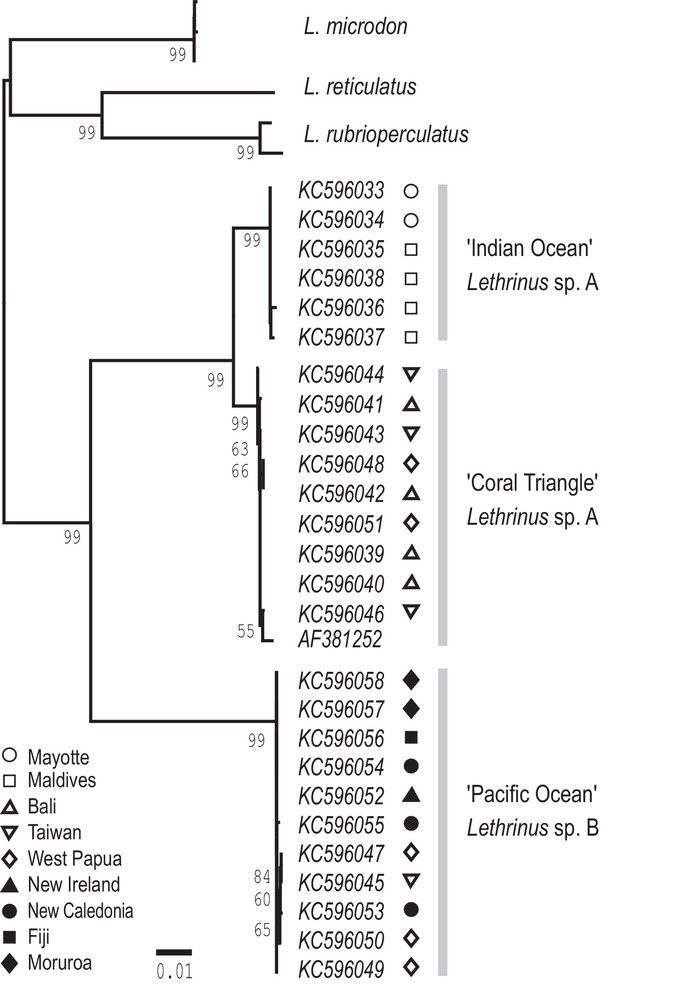
Longface emperor, Lethrinus olivaceus. Phylogenetic tree of mitochondrial lineages reconstructed using RAxML [17], from the cyt-b gene sequences of individuals from the following sampling locations: Bancs Geyser and Zélée, Mayotte (N = 2); Baa atoll, Maldives (N = 4); Kedonganan fish market, Bali (N = 4); Philippines (N = 1: GenBank AF381252 [16]); Kenting fish market, Taiwan (N = 4); Raja Ampat, West Papua (N = 5); Kavieng, New Ireland (N = 1); New Caledonia (N = 3); Tai Levu, Fiji (N = 1); and Moruroa atoll, French Polynesia (N = 2). The tree was rooted by cyt-b sequences of the known closest relatives to L. olivaceus [16], namely L. reticulatus (GenBank AF381253), L. rubrioperculatus (GenBank AF240752, AF381263) and L. microdon (from Bali and the Maldives, and GenBank AF381265 from the Philippines). Branch length proportional to number of substitutions under the GTR + G model. Numbers on branches are ML bootstrap values; those below 50% are not shown.
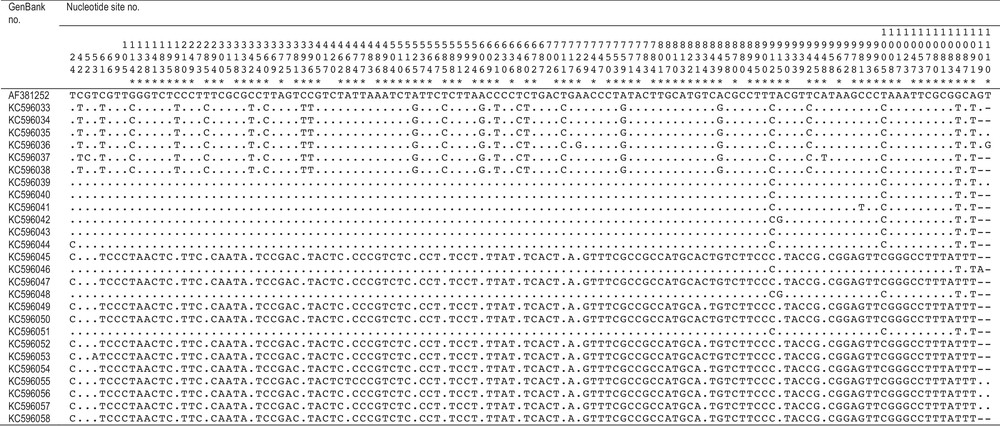
Variable nucleotide sites in cytochrome b gene sequences of longface emperor, Lethrinus olivaceus. Nucleotide sites numbered and mutations marked using Lo Galbo et al.’s sequence from the Philippines (GenBank AF381252) [16] as reference. Asterisk: nucleotide site diagnostic between Lethrinus sp. A and Lethrinus sp. B; dot: nucleotide identical to reference; dash: no data.
4 Discussion
In the only molecular phylogeny of species of the genus Lethrinus published to date [16], the nucleotide divergence at the cytochrome b locus between sister species at the extremity of the tree was comprised between 7% and 14%. The lowest value in this comparison (7%, between L. lentjan and L. ornatus) separates lethrinid species which are morphologically distinct by current taxonomic standards [2]. Using this as a yardstick, the deep branching in the phylogenetic tree (Fig. 4) and the associated nucleotide divergence between the “Indian Ocean-Coral Triangle” and “Pacific Ocean” forms of L. olivaceus (9.5%; greater than 7%) indicate distinct species. These two species are here provisionally designated as Lethrinus sp. A and Lethrinus sp. B, respectively. The distinction of two species is further supported by morphometric differences.
The level of divergence between the “Indian-Ocean” and the ‘Coral-Triangle’ forms of Lethrinus sp. A (1.7%), although much lower than morphologically recognizable species pairs in Lethrinidae, may nevertheless be high enough to compare with Indo-West Pacific species pairs in other phylogenetically well-documented fish families, e.g. Acanthuridae [21], Chaetodontidae [22], and Siganidae [23,24]. Pending further investigation, we here adopt a conservative approach by considering the latter two forms to represent allopatric or parapatric populations of a single species (namely, Lethrinus sp. A). The Indo-West Pacific barrier, which runs from the Indo-Malay Sunda Shelf in the west to the Papuan-Australian Sahul shelf in the east, separates the marine biota of the Indian Ocean and of the Pacific Ocean [25]. Both shelves were exposed to form uninterrupted continental masses west and east of the Indo-Malay-Papua archipelago when the sea level fell during glacial periods, thus restricting the water flow between the two oceans [26]. Pleistocene vicariance on either side of the Indo-West Pacific barrier may account for the genetic differences observed between the ‘Indian Ocean’ lineage and the ‘Coral Triangle’ lineage of Lethrinus sp. A. Nuclear genotyping will be helpful to assess the degree of reproductive isolation between these two forms. Further examination based on time-calibrated molecular phylogenetic analysis will certainly be helpful to address more specifically the evolutionary history of this species.
It has been argued here that the two main clades observed within L. olivaceus correspond to distinct species, Lethrinus sp. A and Lethrinus sp. B. It is now necessary to assign to either of the latter the type specimens of L. olivaceus (MNHN 0000-5741) and L. rostratus (RMNH D443). Depending on the results, it might further be necessary to do so with L. waigiensis (MNHN 0000-9065), and it might also be necessary to collect specimens from the Caroline Islands, the type locality of L. xanthopterus (a nomen nudum) and assign them similarly. Finally, it might also be necessary to examine the type specimens of L. longirostris (BMNH 1865.9.21.6), the last in the list of nominal species synonymised with L. olivaceus [2]. To base the assignment on type locality would be hazardous: the type-localities of both L. olivaceus (Sunda Strait, Indonesia) and L. waigiensis (Waigeo Island, West Papua) lie within the Coral Triangle where both species may occur, according to our genetic evidence. Also, the type locality of L. rostratus has not been clearly indicated by A. Valenciennes, who only mentioned it was “envoyé de Batavia par MM. Kuhl et van Hasselt” [1]. Because of this, and because of possible overlap in the geographic distribution of the two species, nucleotide sequencing the type specimens at the cytochrome b locus is warranted. In Table 1, we propose a molecular-based strategy to rationalize the actions needed for the purpose of revising the taxonomy and nomenclature of species currently under L. olivaceus. Additional nomenclatural problems might arise in case the “Coral-Triangle” form and the ‘Indian Ocean’ form also proved upon further research to be distinct species within Lethrinus sp. A.
Recommended molecular-based strategy to solve the nomenclatural problem posed by the occurrence of two species within L. olivaceus under its current definition [2]. The actions to be done are arranged in a chronological order compatible with page priority regarding species names in [1] and chronological priority otherwise [27].
| Step no. | Action, hypothetic result | Conditional taxonomic decision | Next step |
| 1. | Sequence holotype of L. olivaceus (MNHN 0000-5741) | ||
| 1a. if MNHN 0000-5741 = Lethrinus sp. A (1a) | then Lethrinus sp. A = L. olivaceus | go to 2 | |
| 1b. if MNHN 0000-5741 = Lethrinus sp. B (1b) | then Lethrinus sp. B = L. olivaceus | go to 2 | |
| 2. | Sequence holotype of L. rostratus (RMNH D443) | ||
| 2a. if RMNH D443 = Lethrinus sp. A | then Lethrinus sp. A = L. rostratus | if (1a) true then 3; else 4 | |
| 2b. if RMNH D443 = Lethrinus sp. B | then Lethrinus sp. B = L. rostratus | if (1b) true then 3; else 4 | |
| 3. | – | Maintain synonymy of L. rostratus with L. olivaceus | go to 5 |
| 4. | – | Resurrect L. rostratus | end |
| 5. | Sequence holotype of L. waigiensis (MNHN 0000-9065) | ||
| 5a. if MNHN 0000-9065 = Lethrinus sp. A | then Lethrinus sp. A = L. waigiensis | if (1a) true then 6; else 7 | |
| 5b. if MNHN 0000-9065 = Lethrinus sp. B | then Lethrinussp. B = L. waigiensis | if (1b) true then 6; else 7 | |
| 6. | – | Maintain synonymy of L. waigiensis with L. olivaceus | go to 8 |
| 7. | – | Resurrect L. waigiensis | end |
| 8. | Sequence specimens from the Caroline Islands | ||
| 8a. if Caroline Islands specimens = Lethrinus sp. A | then Lethrinus sp. A = L. xanthopterus | if (1a) true then 9; else 10 | |
| 8b. if Caroline Islands specimens = Lethrinus sp. B | then Lethrinus sp. B = L. xanthopterus | if (1b) true then 9; else 10 | |
| 9. | – | Maintain synonymy of L. xanthopterus with L. olivaceus | go to 11 |
| 10. | Choose neotype for L. xanthopterus | Resurrect L. xanthopterus | end |
| 11. | Sequence holotype of L. longirostris (BMNH 1865.9.21.6) | ||
| 11a. if BMNH 1865.9.21.6 = Lethrinus sp. A | then Lethrinus sp. A = L. longirostris | if (1a) true then 12; else 13 | |
| 11b. if BMNH 1865.9.21.6 = Lethrinus sp. B | then Lethrinus sp. B = L. longirostris | if (1b) true then 12; else 13 | |
| 12. | – | Maintain synonymy of L. longirostris with L. olivaceus | if (1a) true then 14; else 15 |
| 13 | – | Resurrect L. longirostris | end |
| 14. | – | Lethrinus sp. B = undescribed species from the Indo-West Pacific | go to 16 |
| 15. | – | Lethrinus sp. A = undescribed species from the Indian Ocean and the Coral Triangle | go to 16 |
| 16. | – | Describe new species | end |
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
We thank P. Boblin (Secretariat of the Pacific Community, Nouméa), F. Giancarlo (Proyek BioKoR, Kerobokan), H.-C. Ho (National Museum of Marine Biology and Aquarium, Pingtung), and L. Olonde (vendor at the Nouméa fish market) for providing samples. Access to samples from the Mozambique Channel was through the CoPeMay cooperative in Mamoudzou, Mayotte. The fish from Moruroa were collected by our IRD colleagues M. Kulbicki and G. Mou Tham during Cruise ISOTOPE of RV Alis. We thank S. Pedron and A. Collet for help in initiating the current project at IRD Nouméa, C.-K. Huang (M.Sc. student, National Taiwan University, Taipei) for help with the production of sequences, A. Acitino (B.Sc. student at Université de la Nouvelle-Calédonie, Nouméa) for help in measuring head skeletons, D. Ponton(IRD, Nouméa) for advice on the morphometric analyses, P. Béarez, G. Duhamel, Z. Gabsi and P. Pruvost (MNHN, Paris) for help with examining type material, A. Syahailatua and M. Suharsono (LIPI-P2O, Jakarta) for allowing PB to export tissue samples from Indonesia, and S. Andréfouët, L. Scott and the Maldives Marine Research Center for help while doing field work in the Maldives. Access to 19th-century books was through the Biodiversity Heritage Library Internet site (http://www.biodiversitylibrary.org). Designed the study: P.B., W.J.C. Contributed reagents or materials or analysis tools: P.B., W.J.C. Did the experiments: P.B., D.R.H. Analyzed and interpreted the data: P.B., K.E.C., W.J.C., D.R.H. Wrote the paper: P.B., W.J.C. Edited the final version: K.E.C. Financed in part by IRD-UR 227, by the ZoNéCo program (New Caledonia), by LIPI/Ekspedisi Widya Nusantara to West Papua in November-December 2007; by a grant from AIRD/IFB Program “Biodiversité des îles de l’Océan Indien” to S. Andréfouët, by Program CNRS-INEE/TAAF “Iles Eparses” (PhylIP project: P.B.), and by the National Science Council of Taiwan (grants nos. NSC 99-2611-M-002-001-MY2 and NSC 101-2611-M-002-016-MY3 to WJC). Funding for KEC was in part by NSF-0730256.
Appendix 1 Lethrinus spp. Material examined for nucleotide sequence variation at the cytochrome b locus, with sampling details and GenBank (http://www.ncbi.nlm.nih.gov/) sequence accession numbers, and comparative material from GenBank. IRD number allocated by PB to tissue sample preserved in the tissue collection of IRD-UR 227 (Montpellier, France); NTU number allocated by WJC to aliquot preserved at the Marine Phylogenomics Laboratory of NTU-Institute of Oceanography (Taipei, Taiwan). USNM United States Natural History Museum, Washington DC; LIPI-P2O Lembaga Ilmu Pengetahuan Indonesia-Pusat Penelitian Oseanografi, Jakarta; MNHN Museum national d’histoire naturelle, Paris.
| Species, sampling location | Sampling date | Tissue no. | Voucher | GenBank | |
| IRD | NTU | ||||
| Lethrinus microdon | |||||
| Maldives | June 2008 | Lmic 117 | Let110 | – | KC596059 |
| Bali | January 2005 | Z201 | Let151 | – | KC596060 |
| Philippines | – | – | – | USNM 340523 | AF381265 |
| Lethrinus olivaceus | |||||
| Mayotte | April 2011 | PhylIP, L. olivaceus | Let362 | – | KC596033 |
| Mayotte | April 2011 | PhylIP, L. olivaceus | Let363 | – | KC596034 |
| Maldives | June 2008 | 20080621 XL | Let049 | IRD, unregistered (skull) | KC596035 |
| Maldives | June 2008 | Loli 124 | Let117 | – | KC596036 |
| Maldives | June 2008 | 20080621 L | Let128 | IRD, unregistered (skull) | KC596037 |
| Maldives | June 2008 | Loli 83 | Let130 | – | KC596038 |
| Bali | February 2007 | Loli 23 | Let047 | IRD, unregistered (skull) | KC596039 |
| Bali | February 2007 | Loli 24 | Let141 | IRD, unregistered (skull) | KC596040 |
| Bali | February 2007 | Loli 25 | Let142 | IRD, unregistered (skull) | KC596041 |
| Bali | February 2007 | Loli 34 | Let150 | – | KC596042 |
| Taiwan | May 2011 | – | Let382 | 20110520HC004 | KC596043 |
| Taiwan | May 2011 | – | Let384 | 20110506HC006 | KC596044 |
| Taiwan | May 2011 | – | Let385 | 20110513HC010 | KC596045 |
| Taiwan | June 2011 | – | Let431 | – | KC596046 |
| Philippines | - | – | – | USNM 345988 | AF381252 |
| West Papua: Raja Ampat | November 2007 | Gutila 11 | Let050, Let164 | LIPI-P2O, unregistered | KC596047 |
| West Papua: Raja Ampat | November 2007 | Gutila | Let155 | – | KC596048 |
| West Papua: Raja Ampat | November 2007 | Gutila 3 | Let156 | – | KC596049 |
| West Papua: Raja Ampat | November 2007 | Gutila 9 | Let162 | – | KC596050 |
| West Papua: Raja Ampat | November 2007 | Gutila 12 | Let165 | IRD, unregistered (skull) | KC596051 |
| New Ireland: Kavieng | August 2006 | Loli 15 | Let166 | - | KC596052 |
| New Caledonia: N lagoon | April 2007 | Loli 44 | Let171 | IRD, unregistered (skull) | KC596053 |
| New Caledonia: N lagoon | April 2007 | Loli 48 | Let175 | IRD, unregistered (skull) | KC596054 |
| New Caledonia: Chesterfield Is. | August 2011 | – | Let423 | – | KC596055 |
| Fiji: Tai Levu | September 2008 | Loli 70 | Let060 | – | KC596056 |
| Moruroa | July 2006 | Loli M | Let048, Let234 | MNHN 2009-0016 | KC596057 |
| Moruroa | July 2006 | Loli 3 | Let218 | IRD, unregistered (skull) | KC596058 |
| Lethrinus reticulatus | |||||
| Philippines | – | – | – | USNM 340558 | AF381253 |
| Lethrinus rubrioperculatus | |||||
| Western Australia | – | – | – | – | AF240752 |
| Philippines | – | – | – | USNM 346714 | AF381263 |
Appendix 2 Lethrinus olivaceus. Measurements on neurocranium, left pre-maxilla (upper jaw) and left lower jaw (see Fig. 3A), made to the nearest 0.1 mm using a Vernier calliper: cbL, condylo-basal length; dO, orbit diameter; H, neurocranium height; iO, interorbital width; Llj, length of lower jaw; Lpmx, length of premaxilla; pmx, length of toothed part of premaxilla; sw, width of snout where narrowest; w, neurocranium width. The unit for all measurements is the mm.
| Specimen no. | Sampling location | Morphometric variable | ||||||||
| cbL | H | w | sw | iO | dO | Lpmx | pmx | Llj | ||
| Loli 3a | Moruroa | 119.1 | 55.4 | 50.6 | 13.8 | 30.2 | 34.9 | 42.0 | 76.3 | 56.9 |
| Loli 4 | Moruroa | 94.3 | 44.4 | 41.7 | 11.8 | 25.2 | 29.0 | 30.2 | 57.5 | 43.6 |
| Loli 5 | Moruroa | 123.7 | 58.4 | 54.7 | 14.5 | 34.3 | 38.0 | 43.8 | 78.1 | 61.1 |
| Loli 6 | Moruroa | 90.0 | 43.9 | 40.3 | 10.9 | 24.4 | 28.0 | 31.0 | 54.8 | 42.9 |
| Loli 7 | Moruroa | 125.3 | 59.1 | 55.3 | 15.9 | 40.0 | 38.0 | 44.4 | 80.2 | 60.7 |
| Loli 8 | Moruroa | 126.3 | 58.4 | 55.6 | 13.0 | 34.5 | 38.0 | 46.7 | 80.4 | 62.9 |
| Loli 9 | Moruroa | 90.5 | 43.4 | 40.7 | 11.6 | 24.9 | 28.7 | 29.4 | 52.1 | 42.0 |
| Loli 10 | Moruroa | 86.6 | 40.8 | 37.6 | 10.1 | 22.9 | 28.1 | 28.6 | 52.0 | 41.3 |
| Loli 21 | Bali | 84.3 | 38.8 | 36.1 | 10.6 | 27.2 | 25.9 | 28.9 | 54.9 | 42.0 |
| Loli 23a | Bali | 111.9 | 50.6 | 46.4 | 12.1 | 27.8 | 33.1 | 39.6 | 74.0 | 57.9 |
| Loli 24a | Bali | 90.2 | 40.9 | 37.8 | 10.2 | 22.1 | 28.6 | 31.3 | 57.3 | 44.3 |
| Loli 25a | Bali | 81.2 | 36.6 | 35.8 | 8.8 | 19.4 | 27.1 | 27.8 | 50.1 | 39.3 |
| 20080621 La | Maldives | 130.8 | 59.9 | 47.5 | 14.3 | 33.7 | 39.7 | 47.8 | 90.6 | 69.8 |
| Gutila 12a | West Papua | 129.8 | 58.6 | 55.0 | 13.3 | 33.6 | 38.6 | 56.9 | 105.5 | 80.2 |
| Loli 40 | New Caledonia | 99.1 | 47.5 | 44.3 | 11.4 | 26.0 | 29.7 | 35.3 | 63.6 | 47.8 |
| Loli 41 | New Caledonia | 102.2 | 49.0 | 44.6 | 12.3 | 27.3 | 31.0 | 34.8 | 64.2 | 49.3 |
| Loli 42 | New Caledonia | 92.0 | 43.4 | 40.3 | 11.7 | 24.1 | 28.8 | 30.6 | 58.0 | 43.5 |
| Loli 43 | New Caledonia | 88.6 | 41.7 | 38.1 | 10.9 | 22.0 | 28.0 | 29.6 | 55.3 | 41.0 |
| Loli 44a | New Caledonia | 91.0 | 43.5 | 39.8 | 11.1 | 24.0 | 29.0 | 30.4 | 57.0 | 43.4 |
| Loli 45 | New Caledonia | 98.8 | 46.9 | 44.0 | 12.2 | 26.8 | 30.8 | 33.6 | 62.9 | 48.0 |
| Loli 46 | New Caledonia | 97.9 | 47.3 | 43.8 | 12.2 | 25.8 | 29.5 | 33.9 | 61.7 | 47.7 |
| Loli 47 | New Caledonia | 99.6 | 47.9 | 44.1 | 12.1 | 25.8 | 29.5 | 34.0 | 64.3 | 48.7 |
| Loli 48a | New Caledonia | 84.0 | 40.4 | 38.0 | 10.5 | 22.7 | 25.6 | 27.8 | 51.8 | 39.6 |
| Loli 49 | New Caledonia | 100.5 | 47.4 | 44.0 | 12.1 | 26.2 | 30.9 | 34.1 | 64.5 | 46.7 |
| Loli 50 | New Caledonia | 85.1 | 40.3 | 37.1 | 9.3 | 21.7 | 28.4 | 28.7 | 52.5 | 41.1 |
| Loli 51 | New Caledonia | 98.9 | 47.6 | 43.0 | 11.0 | 25.7 | 30.1 | 33.1 | 60.8 | 45.9 |
| Loli 52 | New Caledonia | 95.2 | 46.1 | 41.9 | 10.7 | 25.2 | 29.5 | 31.2 | 59.5 | 42.3 |
| Loli 53 | New Caledonia | 80.9 | 38.4 | 36.8 | 9.7 | 20.8 | 26.6 | 26.9 | 49.4 | 38.0 |
| Loli 54 | New Caledonia | 91.5 | 44.3 | 39.7 | 10.4 | 24.0 | 29.5 | 31.2 | 58.4 | 45.1 |
| Loli 55 | New Caledonia | 89.7 | 42.9 | 39.5 | 10.2 | 22.7 | 27.5 | 30.8 | 56.8 | 43.1 |
| Loli 56 | New Caledonia | 98.8 | 47.2 | 43.0 | 11.3 | 25.3 | 30.5 | 33.4 | 62.8 | 46.8 |
| Loli 57 | New Caledonia | 91.2 | 43.2 | 40.0 | 10.4 | 24.0 | 29.0 | 30.6 | 57.9 | 44.8 |
| Loli 58 | New Caledonia | 90.0 | 44.1 | 40.3 | 9.9 | 23.0 | 29.0 | 29.7 | 56.3 | 43.1 |
| Loli 59 | New Caledonia | 99.4 | 47.8 | 41.4 | 11.5 | 24.8 | 29.3 | 32.0 | 60.4 | 45.8 |
| Loli 60 | New Caledonia | 93.2 | 44.7 | 40.9 | 10.7 | 23.4 | 28.7 | 31.2 | 57.7 | 44.8 |
| Loli 61 | New Caledonia | 95.7 | 45.3 | 42.2 | 10.8 | 24.8 | 29.2 | 32.2 | 58.0 | 47.0 |
a also sequenced at the cytochrome b locus.


