1 Introduction
Wheat is considered one of the most widely cultivated and consumed food crops in the world. Cultivated wheats are either hexaploid (Triticum aestivum, AABBDD, 2n = 6x) or tetraploid (Triticum durum or Triticum turgidum subsp. durum, AABB, 2n = 4x). This genomic complexity makes it difficult to accurately sequence and assemble the entire nuclear genome. The draft genome of the A-genome progenitor species (Triticum urartu, AA) has been assembled and assigned as the diploid reference for further analysis of polyploid nuclear wheat genomes [1]. The available reference plastid genome of the hexaploid “Chinese Spring” cultivar was completed by Sanger sequencing of a set of cloned restriction fragments that covered the entire genome [2]. As part of a project to examine plastid single nucleotide polymorphisms (SNPs) among nine wheat cultivars from Egypt, we sequenced the complete genome and discovered that the published wheat genome contains contaminated sequence from the rice plastid genome. In this paper, we characterize the corrected plastid genome sequence for one cultivar of wheat from Egypt.
2 Materials and methods
2.1 DNA Isolation, genome sequencing and mapping of reads to reference plastid genome
Total genomic DNA was extracted from leaf tissues (∼1 g) of 14-day-old etiolated seedlings of one hexaploid wheat cultivar (Giza 168, Delta, Egypt) using the modified procedure of [3]. Purified total genomic DNA was sent to Beijing Genomics Institute (BGI), Shenzhen, China for sequencing using the Illumina HiSeq 2000 platform. Thirty million 100-bp paired-end reads were generated from a sequencing library with 500-bp inserts. Adapter sequences in reads of the raw data were deleted, and reads with 50% low quality bases (quality value ≤ 5) or more were removed. The remaining sequences were mapped to the published wheat plastid genome (accession number NC_002762) using CLC Genomics Workbench (version 3.0, http://www.clcbio.com/usermanuals). The raw sequence reads (SRA XXXX) and the assembled and annotated plastid genome sequence of cultivar Giza 168 (accession number KJ592713) were deposited in NCBI. Annotation of the plastid genome was performed using DOGMA [4] supplemented with tRNAscan (http://lowelab.ucsc.edu/tRNAscan-SE/) and ARAGORN (http://mbioserv2.mbioekol.lu.se/ARAGORN) for tRNAs. A circular genome map was constructed with GenomeVx [5].
3 Results and discussion
Raw reads were mapped to the reference wheat plastid genome (accession number NC_002762). The number of reads mapped was 1,195,172, which represents 1.1% of the total reads. The read depth averaged 1,229X coverage across the genome. The assembled plastid genome of the Giza 168 wheat cultivar is 133,873 bp, which is 672-bp smaller than the published genome for the “Chinese Spring” cultivar [2]. Mapping of the Giza 168 genome to the “Chinese Spring” genome identified three DNA sequences of 332, 131 and 131 bp that are absent from the Giza 168 cultivar (Fig. 1). Other shorter sequences were also found only in the published reference plastid genome. The 332, 131 and 131 bp DNA sequences are located at positions 6,164-6,495, 83,918-84,048 and 130,960-131,090 of the reference genome, respectively. Alignment of these two genomes generated gaps within the plastid genome sequence of the Giza 168 cultivar (Fig. 1). BlastN analyses of the three extra sequences from the wheat reference plastid genome to the NCBI database indicated 100% sequence identity of these fragments to plastid genome of rice (Oryza sativa, Japonica group) as well as the published “Chinese Spring” reference wheat genome (Table 1). The next best Blast hits were to another rice species, Oryza rufipogon with 99% identity. Blast hits to plastid genomes of other cereals were not detected. To further confirm that the published wheat plastid genome sequence is contaminated with rice sequence, the sequences from the wheat GZ168 cultivar flanking the three gaps were blasted to the NCBI database (Table 2). These sequences are located in the plastid genome of cultivar GZ168 between 5,957-6,288 (332 bp), 83,385-83,515 (131 bp) and 130,192-130,322 (131 bp) bases (Fig. 1). The results indicated 100% sequence identity to plastid genome sequences of hexaploid wheat and other members of the Triticeae, while no Blast hits were detected to any of the rice plastid genome sequences available in the GenBank. Annotation of the corrected wheat plastid genome confirmed the gene content and order from the published genome (Fig. 2).
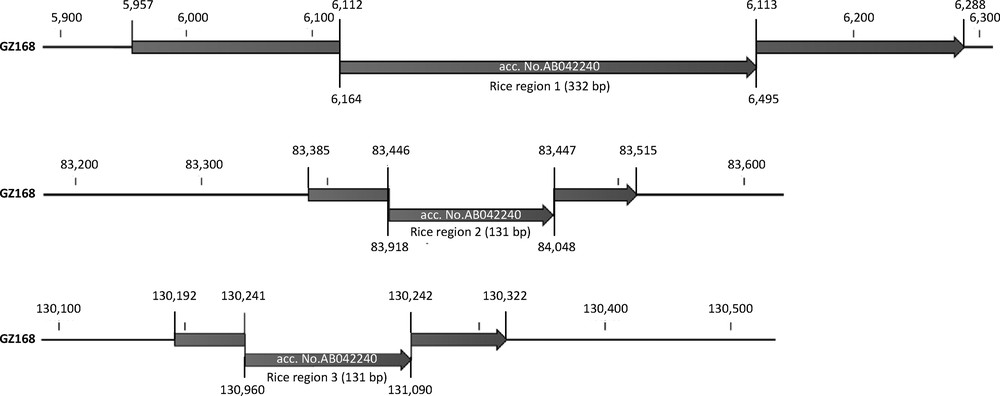
Extra sequences present in the published wheat plastid genome (accession number NC_002762 = AB042240, 134,545 bp) missing in the Egyptian wheat cultivar (GZ168, accession number KJ592713, 133,873 bp). Numbers above and below maps indicate coordinates in KJ592713 and NC_002762, respectively.
BLAST analysis of the three rice cp regions within the wheat cp genome of Chinese Spring cultivar (acc. No. AB042240).
| Description | Max score | Total score | Quary cover | E value | Identity | Accession |
| Rice region 1 (T. aestivum published chloroplast genome of Chinese Spring cv., acc. No. AB042240, bases 6,164-6,495) | ||||||
| Oryza sativa Japonica group cultivar Nipponbare voucher AC01-1001045 chloroplast, complete genome | 614 | 614 | 100% | 2e–172 | 100% | GU592207 |
| Oryza sativa Japonica group isolate PA64S chloroplast, complete genome | 614 | 614 | 100% | 2e–172 | 100% | AY522331 |
| Oryza sativa Japonica group cultivar Nipponbare chloroplast, complete genome | 614 | 614 | 100% | 2e–172 | 100% | AY522330 |
| Oryza sativa Japonica group chloroplast genome | 614 | 614 | 100% | 2e–172 | 100% | X15901 |
| Triticum aestivum chloroplast DNA, complete genome | 614 | 614 | 100% | 2e–172 | 100% | AB042240 |
| Oryza rufipogon cultivar DongXiang chloroplast, complete genome | 608 | 608 | 100% | 2e–171 | 99% | KF562709 |
| Oryza rufipogon chloroplast, complete genome | 608 | 608 | 100% | 2e–171 | 99% | JN005832 |
| Rice region 2 (T. aestivum published chloroplast genome of Chinese Spring cv., acc. No. AB042240, bases 83,918-84,048) | ||||||
| Oryza sativa Japonica group cultivar Nipponbare voucher AC01-1001045 chloroplast, complete genome | 241 | 482 | 100% | 1e–60 | 100% | GU592207 |
| Oryza sativa Japonica group isolate PA64S chloroplast, complete genome | 241 | 482 | 100% | 1e–60 | 100% | AY522331 |
| Oryza sativa Japonica group cultivar Nipponbare chloroplast, complete genome | 241 | 482 | 100% | 1e–60 | 100% | AY522330 |
| Oryza sativa Japonica group chloroplast genome | 241 | 482 | 100% | 1e–60 | 100% | X15901 |
| Triticum aestivum chloroplast DNA, complete genome | 241 | 482 | 100% | 1e–60 | 100% | AB042240 |
| Oryza rufipogon cultivar DongXiang chloroplast, complete genome | 241 | 482 | 100% | 1e–60 | 100% | KF562709 |
| Oryza rufipogon chloroplast, complete genome | 241 | 482 | 100% | 1e–60 | 100% | JN005832 |
| Rice region 3 (T. aestivum published chloroplast genome of Chinase Spring cv., acc. No. AB042240, bases 130,960-131,090) | ||||||
| Oryza sativa Japonica group cultivar Nipponbare voucher AC01-1001045 chloroplast, complete genome | 241 | 486 | 100% | 4e–61 | 100% | GU592207 |
| Oryza sativa Japonica group isolate PA64S chloroplast, complete genome | 243 | 486 | 100% | 4e–61 | 100% | AY522331 |
| Oryza sativa Japonica group cultivar Nipponbare chloroplast, complete genome | 243 | 486 | 100% | 4e–61 | 100% | AY522330 |
| Oryza sativa Japonica group chloroplast genome | 243 | 486 | 100% | 4e–61 | 100% | X15901 |
| Triticum aestivum chloroplast DNA, complete genome | 243 | 486 | 100% | 4e–61 | 100% | AB042240 |
| Oryza rufipogon cultivar DongXiang chloroplast, complete genome | 243 | 486 | 100% | 4e–61 | 100% | KF562709 |
| Oryza rufipogon chloroplast, complete genome | 243 | 486 | 100% | 4e–61 | 100% | JN005832 |
BLAST analysis of regions in the wheat cp genome of GZ168 cultivar (acc. No. KJ592713) flanking rice cp regions.
| Description | Max score | Total score | Quary cover | E value | Identity | Accession |
| Flanking region 1 (T. aestivum chloroplast genome of GZ168 cv., acc. No. KJ592713, bases 5,957-6,288) | ||||||
| Triticum aestivum chloroplast, complete genome | 614 | 614 | 100% | 2e–172 | 100% | KC912694 |
| Aegilops speltoides isolate SPE0661 chloroplast, complete genome | 608 | 608 | 100% | 9e–171 | 99% | JQ740834 |
| Triticum urartu chloroplast, complete genome | 608 | 608 | 100% | 9e–171 | 99% | KC912693 |
| Triticum monococcum subsp. aegilopoides, complete genome | 608 | 608 | 100% | 9e–171 | 99% | KC912692 |
| Triticum monococcum chloroplast, complete genome | 608 | 608 | 100% | 9e–171 | 94% | KC912690 |
| Secale cereale chloroplast, complete genome | 603 | 608 | 100% | 9e–169 | 89% | KC912691 |
| Aegilops tauschii chloroplast, complete genome | 507 | 507 | 100% | 9e–140 | 89% | JQ754651 |
| Hordeum vulgare subsp. spontaneum chloroplast, complete genome | 411 | 411 | 100% | 9e–111 | 89% | KC912689 |
| Flanking region 2 (T. aestivum chloroplast genome of GZ168 cv., acc. No. KJ592713, bases 83,835-83,515) | ||||||
| Triticum aestivum chloroplast, complete genome | 243 | 243 | 100% | 4e–61 | 100% | KC912694 |
| Aegilops speltoides isolate SPE0661 chloroplast, complete genome | 243 | 243 | 100% | 4e–61 | 100% | JQ740834 |
| Secale cereale chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912691 |
| Hordeum vulgare subsp. spontaneum chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912689 |
| Hordeum vulgare subsp. spontaneum chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912688 |
| Hordeum vulgare subsp. vulgare chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912687 |
| Aegilops tauschii chloroplast, complete genome | 231 | 463 | 100% | 8e–58 | 98% | EF115541 |
| Flanking region 3 (T. aestivum chloroplast genome of GZ169 cv., acc. No. KJ592713, bases 130,192-130,322) | ||||||
| Aegilops speltoides isolate SPE0661 chloroplast, complete genome | 243 | 243 | 100% | 4e–61 | 100% | JQ740834 |
| Triticum aestivum chloroplast, complete genome | 243 | 243 | 100% | 4e–61 | 100% | KC912694 |
| Secale cereale chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912691 |
| Hordeum vulgare subsp. spontaneum chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912689 |
| Hordeum vulgare subsp. spontaneum chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912688 |
| Hordeum vulgare subsp. vulgare chloroplast, complete genome | 231 | 231 | 100% | 8e–58 | 98% | KC912687 |
| Aegilops tauschii chloroplast, complete genome | 231 | 463 | 100% | 8e–58 | 98% | EF115541 |
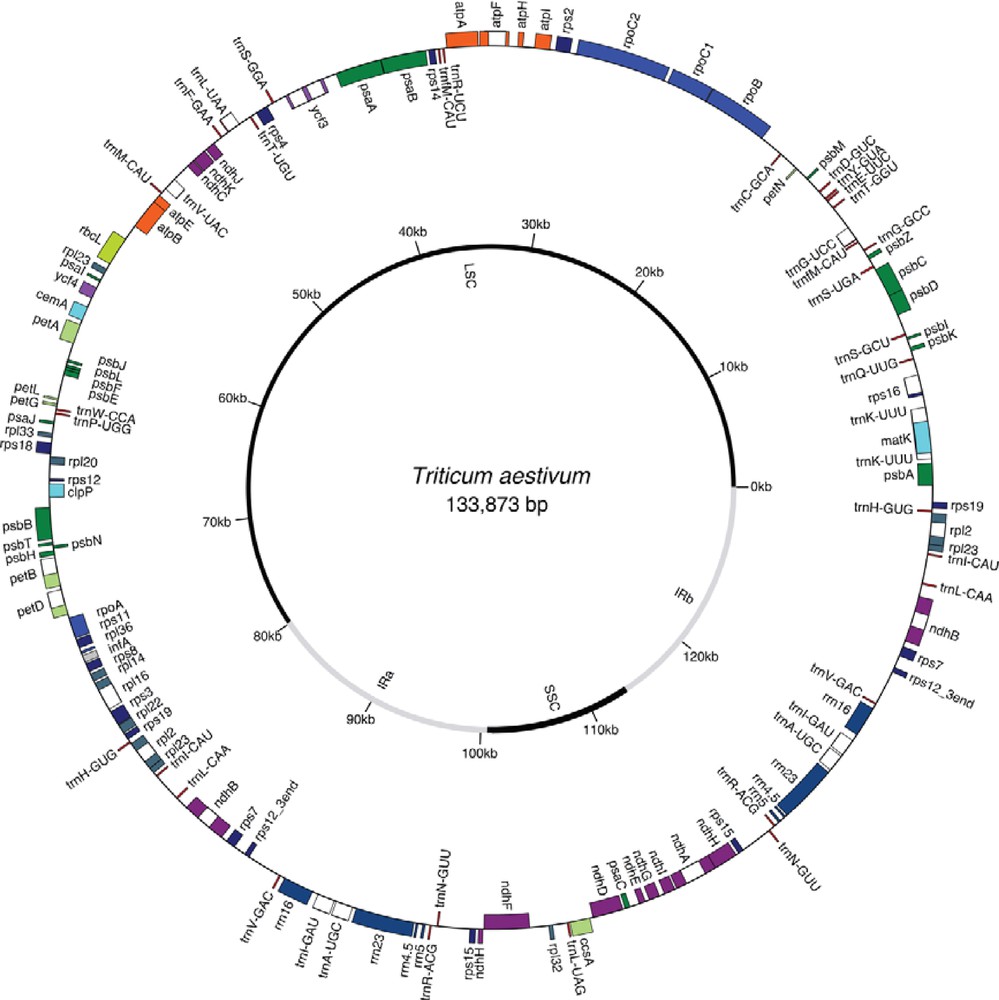
(Color online.) Circular plastid genome map of the GZ168 Egyptian wheat plastid genome (accession number KJ592713). Circle shows gene content with genes outside and inside the ring transcribed counterclockwise and clockwise, respectively.
The cause of the contamination of the wheat plastid genome sequence with rice DNA is unknown, however, other cases of errors in published plastid genome sequences have been detected in the past. For example, in the plastid genome of tobacco [6], sequencing errors of 119 bp were reported seven years after it was published [7]. In the case of tobacco, 90 bp were missed because a small AluI restriction fragment was missed in the cloning and sequencing strategy. One possible explanation for the error in the previously published wheat plastid genome is that the genome was assembled using the rice genome as a reference, and after completion, the entire rice sequences were not removed. Another possibility is that during the cloning of the wheat plastid genome, there was contamination of rice DNA. No matter what the explanation is, we recommend that the corrected wheat genome sequence (accession number KJ592713) be used in all future studies.
Acknowledgements
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under Grant No. (18-3-1432/HiCi). The authors, therefore, acknowledge with thanks DSR technical and financial support. We also thank the Texas Advanced Computing Center (TACC) at the University of Texas, USA for access to supercomputers.


