1 Introduction
Cryptic species, or distinct species described as a single one because of morphological similarity, are common [1], particularly in the marine environment [2]. The majority of cryptic species was discovered during the last decade thanks to the use of molecular methods for species identification, in particular when using mitochondrial DNA-based markers [3,4]. For example, Hoareau et al. [5] investigated the COI and 16S genes in 10 species of brittle star and found seven new lineages, six of which fulfill the criteria to be biological species.
In echinoderms, several examples are known, such as the crinoids Tropiometra carinata [6], Promachocrinus kerguelensis [7,8], Cenolia sp. [7], the sea urchin Echinocardium cordatum [8,9], the sea stars Leptasterias hexactis [10,11], Linkia sp. [12], Patiriella pseudoexigua [13], Parvulastra exigua [14], Cryptasterina pentagona and Cryptasterina hystera [15], the brittle stars Amphipholis squamata [16–18], Ophiocoma erinaceus [19], Ophiothrix fragilis and Ophiothrix quinquemaculata [20–22], Astrotoma agassizii [23,24], and Acrocnida brachiata [25,26], amongst others.
The brittle star Ophioderma longicauda (Bruzelius, 1805) was known as a common Atlanto-Mediterranean gonochoric species [27] reproducing via a lecithotrophic vitellaria larva [28,29]. However, the discovery of brooding individuals [30] suggested that it is actually a species complex composed of six mitochondrial lineages [31]. The lineage L1 at the western Mediterranean French coast reproduces once a year [29] via lecithotrophic vitellaria larvae [28], whereas lineages L2–L3–L4, observed in the Eastern Mediterranean, are also gonochoric, but brood their offspring [30]. Lineages L5 and L6 are expected to reproduce via lecithotrophic larvae as well, but no observation of their reproduction is available yet. The broadcast spawning lineage L1 is found in sympatry with the brooding lineages L2-L3-L4 in the eastern Mediterranean. Brooding females have been reported in the eastern Mediterranean at the end of May/beginning of June [30], whereas the reproduction of lineage L1 was reported at the beginning of July in the western Mediterranean [28].
In the present study, to infer whether there is potential gene flow between brooding and broadcasting lineages, we compared the “reproductive state” of L1 and L3 specimens occurring in sympatry for one month, in May 2012. In addition, we observed the development of brooded juveniles and made inferences about the fertilization process. Furthermore, we investigated morphological characters (maximal disc size, status of radial shields) and bathymetric distribution of both lineages. Finally, to complement the analysis of the mitochondrial marker COI, we analyzed a codominant nuclear marker (thus independent of the mitochondrial marker), which provides another way of testing reproductive isolation between brooders and broadcasters.
2 Methods
2.1 Sampling
Specimens of O. longicauda from lineages L1 and L3 were collected in six locations of Crete during the month of May 2012 (Fig. 1). The sites of collection on the North coast of Crete included: Lygaria Port (LyP) (N 35°23′57.56′′/E 25°01′41.24′′), Lygaria (Lyg) (N 35°24′08.37′′/E 25°02′02.62′′), Gouves (Gou) (N 35° 20′ 13.61′′/E 25° 18′ 18.85′′) and Agios Nikolaos (AgN) (N 35°11′40.84′′/E 25°43′2.37′′). The sites on the southern coast included: Agios Pavlos (AgP) (N 35°06′06.73′/E 24°33′47.82′′) and Ierapetra (Ier) (N 35° 0′ 13.14′′/E 25° 44′ 5.45′′).
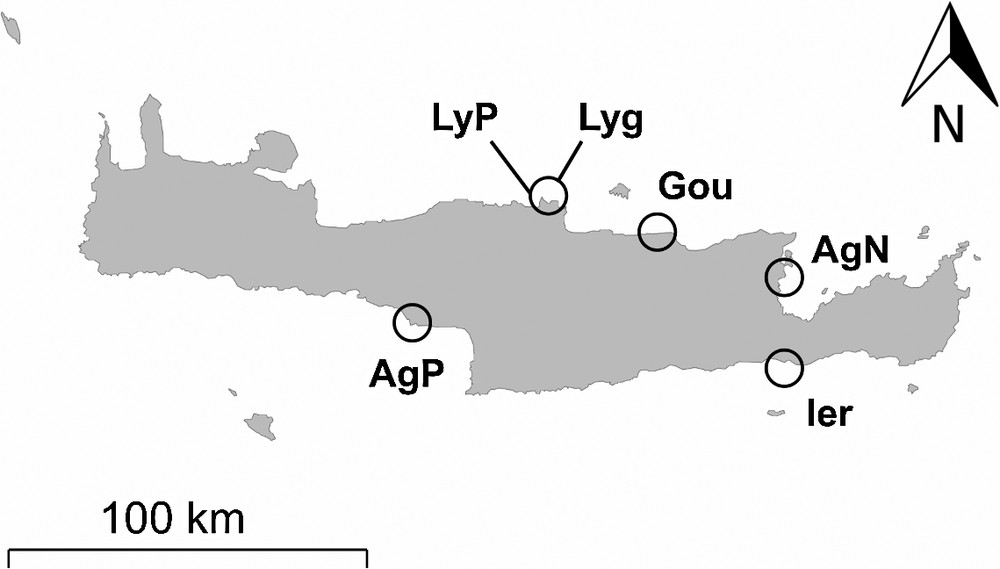
Map of Crete (Greece) with the six localities where individuals of Ophioderma longicauda broadcast spawning lineage L1 and brooding lineage L3 were sampled. LyP: Lygaria Port; Lyg: Lygaria; Gou: Gouves; AgN: Agios Nikolaos; AgP: Agios Pavlos; Ier: Ierapetra.
2.2 DNA extraction, COI sequencing and analyses
Each specimen collected (n = 169) was genotyped to determine its mitochondrial lineage. DNA extractions and PCRs targeting a portion of the COI gene were performed as previously described [30,31]. PCR products were sent for sequencing to a private company (LGC genomics, Berlin, Germany). Sequences were manually edited with the software BioEdit, aligned in the program Seaview [32] with MUSCLE v3.8.31 algorithm [33] and manually improved. Number of haplotypes and haplotype diversity were investigated with DNAsp v5 software [34].
2.3 i51 Genotyping and analyses
The intronic codominant marker (EPIC) i51 [35,36] was used in this study for the first time. PCRs were performed with the primers OL51F2: 5′CCA TAT ATT TAT GTA TTC CCG TGT 3′ and OL51R1: 5′TCA CAG CCA CTT CAT GCT GC AG 3′ in a final volume of 20 μl with the following final reagent concentrations: 1 × of Go Taq flexi buffer, 1.5 mM of MgCl2, 0.2 mM of dNTPs, 0.2 μM of each primer, 0.1 U of Taq Polymerase (Flexi Go Taq, Promega), 1 μL of DNA template diluted ten times and sterile water to complete up to 20 μL. The PCR cycling parameters were as follows: a first step of denaturation at 95 °C 3 min, followed by 35 cycles of 30 s of denaturation at 95 °C, 30 s of annealing at 52 °C, 30 s of elongation at 72 °C and a final step of elongation at 72 °C for 3 min i51 is a codominant marker displaying length heterogeneity with pure microsatellite repetitions of four bases (CCAA motif). A new method using high-resolution agarose electrophoresis was developed for i51 genotyping. It allowed reading the genotype of each individual on agarose gels. Correct amplification of DNA samples was first verified by migrating the PCR products on a regular agarose 2% TBE 1X gel stained with ethidium bromide. Then, a 3.5% high-resolution agarose gel (Sigma-Aldrich) was performed; samples were loaded and migrated during 2.5 h at 120 V. The loaded volume of PCR product was adapted for each individual according to the control gel to optimize resolution. This technique permitted to discriminate homozygous from heterozygous individuals and to differentiate alleles differing by 4 bp or higher multiples of 4 bp. For each gel, one sequenced individual was loaded as a reference for sizing the alleles in two lanes (lanes 7 and 19), as well as the DNA ladder (25 bp DNA step ladder, Promega) in three lanes (on lanes 2, 13 and 25, for a total of 26 lanes). Ethidium bromide staining was done at the end of migration by performing an ethidium bromide bath during 10 min Pictures of the gels were taken with the gel doc (BioRad) and genotypes were determined using the software Quantity One (BioRad).
2.4 Morphological, reproductive and developmental observations
For each specimen collected, the disc diameter (DD) was measured and the status of the radial shields was investigated (covered with granules or naked). Twenty-three individuals of L1 and 34 individuals of L3 were dissected and the status of the gonads was recorded (full, brooding or spawned) to infer the reproductive status of each lineage during the month of May. In addition, specimens from both lineages were preserved for gonad histology. Entire discs were fixed in EtOH 70% for 24 h, dehydrated in EtOH 95% and embedded in araldite resin by successive baths of acetone 100%, acetone 75%–araldite 25% and araldite 100%. Serial sections were stained with toluidine blue and observed with a Leica DM 2500 microscope.
Due to the stress of collection, brooding females released juveniles at different developmental stages. Juveniles were maintained in filtered seawater in glass Petri dishes at ambient temperature, and water was renewed every day. The development of juveniles was monitored and pictures from live specimens were taken at different developmental stages. In addition, some juveniles were preserved in EtOH 95% and prepared for scanning electron microscopy.
3 Results
3.1 Genetic analyses – COI and i51
We obtained a total of 169 COI sequences of 491 bp, 60 sequences belonging to L1 and 109 sequences belonging to L3. A higher number of haplotypes was found in L1 compared to L3 (29 vs. 20; Table 1), and the average haplotype diversity per population was 65% higher in L1 compared to L3 (Table 1).
Comparative summary of morphological characters (disc diameter and status of radial shields) and genetic data (mitochondrial marker COI and nuclear marker i51) in the broadcasting lineage L1 and the brooding lineage L3 of O. longicauda.
| L1 | L3 | |
| N individuals | 60 | 109 |
| N populations | 5 | 5 |
| COI haplotypes (total) | 29 | 20 |
| COI haplotypes (mean per pop) | 8.6 | 5 |
| Hd (mean per pop) | 0.918 | 0.554 |
| i51 alleles (total) | 12 | 1 |
| Radial shields (covered/naked) | 60/0 | 2/107 |
| Disc diameter (mean) | 15.3 mm | 12.3 mm |
The 169 individuals were genotyped for the i51 marker. Lineage L1 displayed 12 alleles for this marker, each 4 bp intermediate being found between alleles 081 bp and 133 bp, except the alleles 101 bp and 113 bp. In contrast, each L3 individual genotyped was homozygous with the allele 081 bp, indicating that i51 was monomorphic in this brooding lineage (Table 1) and constitutes a semi-diagnostic marker of these two lineages.
3.2 Morphology, reproduction and development
3.2.1 Morphological and habitat differences
The radial shields were almost always naked for the L3 specimens (Table 1) and always covered with granules for the L1 specimens (Table 1). Although size distributions were overlapping, mean of disc diameters (DD) differed significantly between L1 and L3 (Student's t test; P < 0.001), with L1 displaying larger specimens than L3 in Crete (L1: mean = 15.3 mm; max = 25 mm; L3: mean = 12.3 mm; max = 21 mm) (Table 1, Fig. 2). Bathymetric distribution differed between L1 and L3 (Fisher exact test, P < 0.001): we found only 15 specimens of L1 and 157 specimens of L3 between 1 m and 3 m depth (including the previously sampled populations of Elounda and Rhodes; see Boissin et al., 2011), whereas we found 48 specimens of L3 and 60 specimens of L1 between 3 m and 12 m
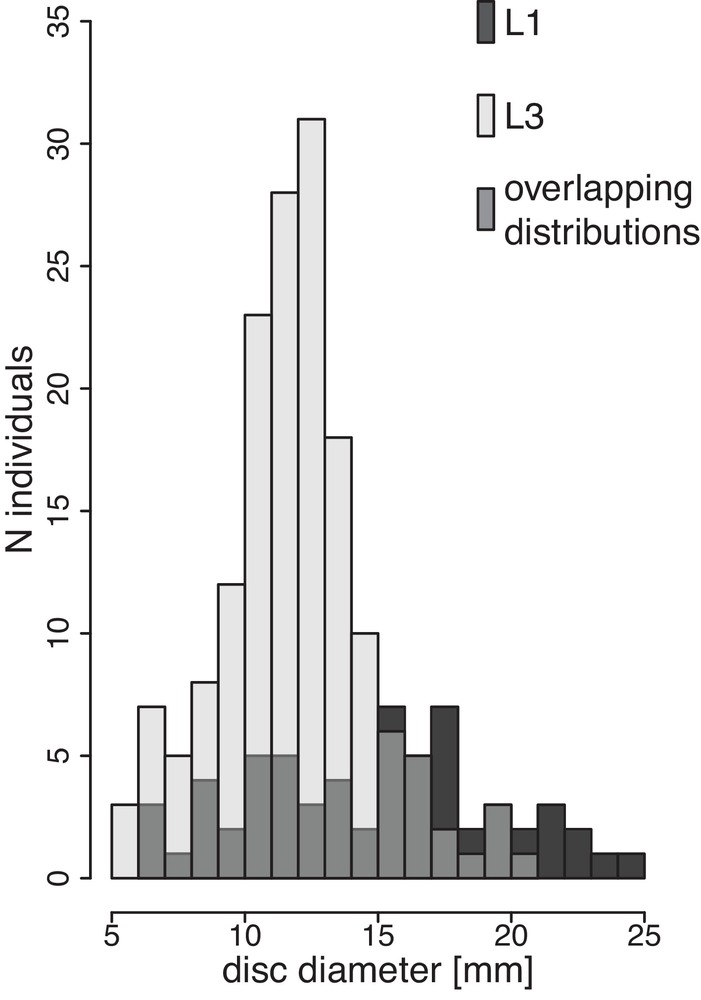
Size distributions between broadcast spawning (L1, n = 60) and brooding (L3, n = 109) genetic lineages. All individuals were sampled in Crete (Greece).
3.2.2 Dissections and histology
3.2.2.1 Lineage L1
Among the 23 dissected individuals genotyped as L1, 16 females and seven males were found, each with visible gonads (Table 2). All observed sections of females displayed ovaries full of late-vitellogenic oocytes (Fig. 3E andF) indicating that the gonads were almost mature. Depending on the section, the number of oocytes per ovary varied between three and 14. No males could be preserved for histology due to their low number. Fertilization trials in aquarium were performed once a week from the beginning of May to the beginning of June as described in [37], but all trials failed, suggesting that L1 individuals were not mature to spawn yet. In addition, we performed dissections of female (Fig. 3A, C, D) and male (Fig. 3B) L1 individuals from Marseilles (France) collected on 13 June 2012. Each specimen displayed full gonads.
Inference of the lineage and the reproductive status of O. longicauda individuals collected from 3 May to 29 May 2012 at five locations in Crete (Greece).
| Observation | Genetic lineage | Conclusion |
| 24 individuals with juveniles | 24 individuals of L3 | Brooding females |
| 16 individuals with eggs | 16 individuals of L1 | Spawning females |
| 11 male individuals with full gonads | 7 individuals of L1 4 individuals of L3 |
Spawning males Late “brooding” males |
| 6 “empty” individuals | 6 individuals of L3 | “brooding” males post reproduction |
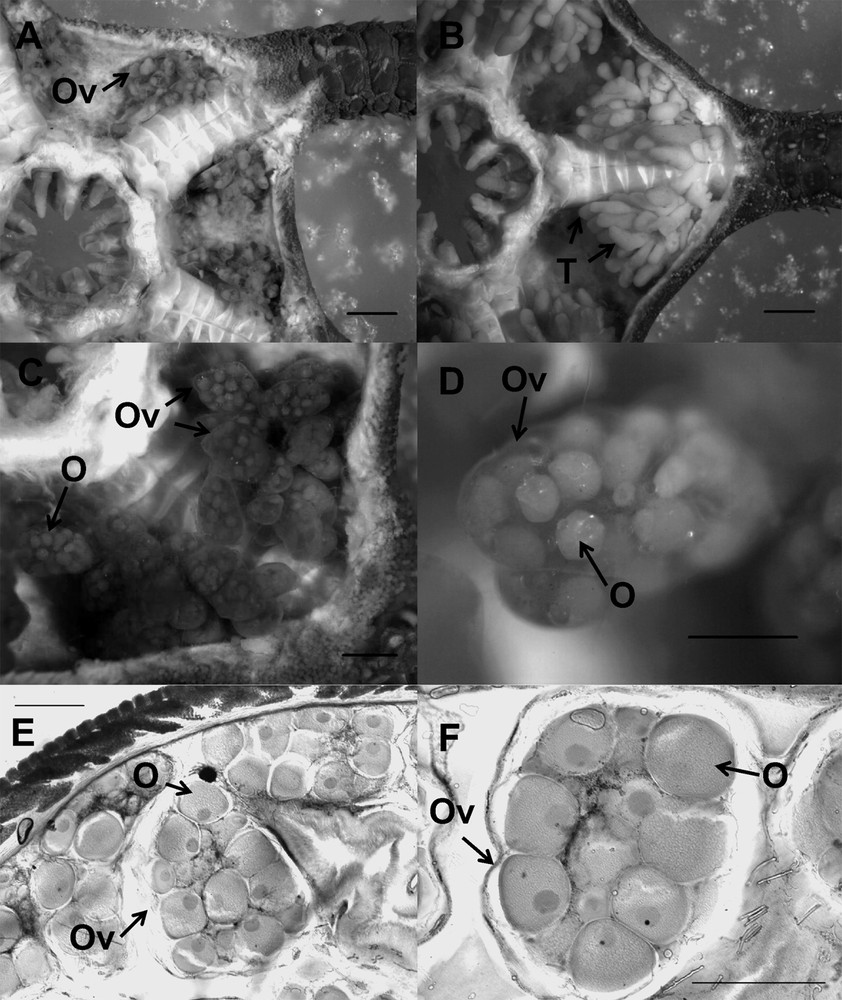
Ophioderma longicauda, broadcast spawning lineage L1. A, C, D: female with the aboral surface of the disc removed to view the ovaries (Ov) containing oocytes (O). B: male with the aboral surface of the disc removed to view the testes (T). E, F: histological sections of ovaries (Ov) containing oocytes (O). Scale bars: A, B: 2 mm; C: 1 mm; D–F: 500 μm.
3.2.2.2 Lineage L3
Among the 34 dissected individuals genotyped as L3, 24 females and 10 males were found (Table 2). Six males displayed spent gonads and four males displayed full gonads, suggesting that the reproduction just occurred for the six individuals and was about to for the four other individuals. Each female was brooding embryos (Fig. 4A) at different developmental stages (Fig. 4B–H), depending on the sampling date (Gastrula for females collected the 3 May, juveniles at different stages for the other sampling dates). Immediately after the first sampling on 3 May and due to the stress of collection, brooding females released numerous embryos at the gastrula stage, indicating that fertilization indeed occurred just a few hours ago (Fig. 4B). Each brooding female found after this date released juveniles at more advanced developmental stages, indicating that the reproduction temporal window lasted only a few days at the population level (synchrony) and even among populations from northern and southern Crete. Furthermore, each juvenile released by a single female displayed the same developmental stage, suggesting that there was a single fertilization event for each female. For lineage L3, development was followed in Petri dishes every day, and no larval stage was observed between the gastrula and the juvenile stage. The development of embryos first consisted of a rearrangement of cells, as the gastrula diameter and the disc diameter of early juveniles were very similar (380 μm, Fig. 4B, 4E). After reaching a certain development stage, the size of the juvenile increased as the disc diameter and the arms grew in length. As the arms were growing, new pairs of tube feet were successively added to the arm, allowing the juveniles to move very early in their development, even while still being brooded by the mother. During growth, the juveniles also developed disc and arm spines (Fig. 4F). Each embryo/juvenile released developed properly without food supply, supporting the hypothesis that ophiuroids are able to absorb dissolved organic matter through the tube feet [38,39].
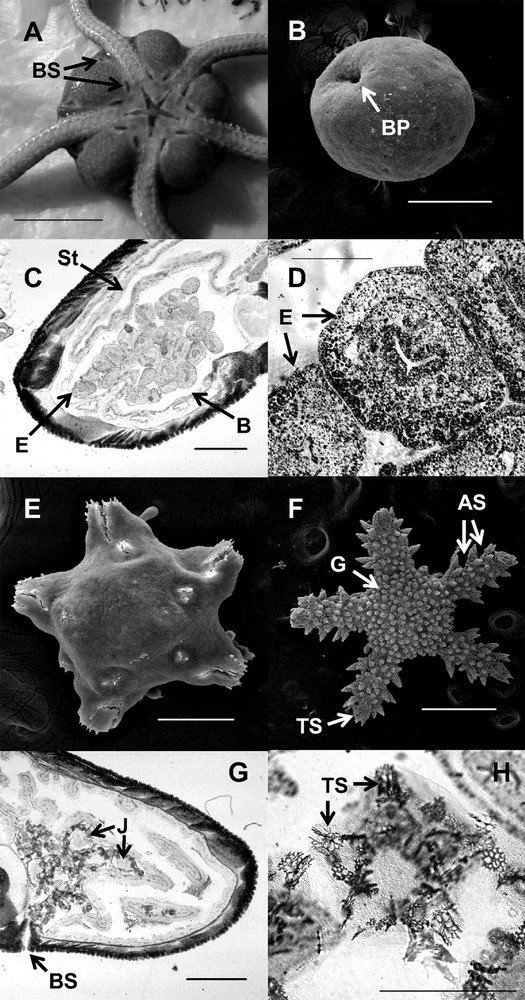
Ophioderma longicauda, brooding lineage L3. A: ventral overview of a brooding female; the gonads are located inside the disc in gonadal chambers (bursae) along both sides of each arm, opening through two bursal slits (BS) each. B: gastrula with blastopore (BP) (SEM). C: cross-section of disc of a brooding female with embryos (E); bursal wall (B); stomach (St) D: embryo (E). E: juvenile dissected from a bursa, arms with one free segment and the terminal plate, the first disc granules just forming. F: larger juvenile, dissected from a bursa, probably almost ready to hatch, arms with three segments (AS) and the terminal plate (TP), disc and arms covered with granules (G). G: cross-section of disc of a brooding female with developed juveniles (J), about to be expelled through the bursal slit (BS). H: mature embryo present in female bursa, probably almost ready to hatch; terminal plate (TP). Scale bars: A: 1 cm; B: 200 μm; C: 1 mm; D, E: 200 μm; F: 500 μm; G: 1 mm; H: 500 μm.
4 Discussion
4.1 Morphological differences
The status of the radial shields was previously investigated by Stöhr et al. [30], who also noticed a tendency for the brooders to display naked radial shields, but this characteristic was not present in every brooding specimen. In contrast, the naked character versus covered radial shields appeared to be diagnostic of the lineage (L1 versus L3) in Crete. Further evidence is needed to determine to what extent this character is reliable on a larger geographic scale and if it is applicable to other brooding lineages L2 and L4. The size distributions of disc diameters overlapped, but very large individuals can be relatively safely diagnosed as belonging to L1. When comparing closely related species in marine invertebrates, the brooding ones are generally smaller than the ones with larvae [40,41]. O. longicauda follows this rule, although size differences are moderate. In this species complex, brooding is most likely the derived character [30], but it is still unclear whether reduced size preceded brooding or not.
4.2 Habitat difference to explain L1 distribution
Although we have to consider habitat differences with caution due to the relatively low number of sampling stations, L1 individuals appeared less abundant than L3 at the shallowest depths (1–3 m). To our current knowledge, L1 individuals are less abundant in the eastern Mediterranean than in the western part, where they largely dominate [31], although this may be an artefact because the majority of samples were collected at shallow depths. One possible explanation is that L3 individuals are more adapted to elevated temperatures than L1, as mean temperatures are more elevated in the eastern basin [42]. Indeed, a thermotolerance experiment conducted with L1 and L3 showed that L3 survival was not significantly affected by elevated temperatures contrary to L1 [43]. Another possibility is that L1 might suffer from the high UV penetration in the eastern basin, especially during the larval stage, as seen in several marine ectotherms [44,45]. As the waters from the eastern Mediterranean basin are known to be very oligotrophic [46], UV rays penetrate deeper into the water column. During the reproduction period, the larvae of L1 may be more exposed to UV radiation as they are expected to swim close to the surface [47]. They may thus display a lower recruitment compared to brooded individuals that are less exposed to UV due to their strictly benthic life, and their likely habit, once released, of hiding under rocks, as do adults. UV also certainly affects adults [45], but echinoderms can develop avoidance mechanisms [48], particularly some brittle stars that live under rocks during the day and are active during the night [49]. Nevertheless, it is possible that L1 is not less abundant in the eastern basin and that this observation is only a sampling artefact, L1 being as abundant but living deeper. In this case, the fact that L1 lives deeper in the eastern than in the western Mediterranean might also be explained by the UV hypothesis.
4.3 Reproduction strategies and reproductive isolation between L1 and L3
The reproduction of O. longicauda was previously described as occurring once a year via lecithotrophic larvae in early July [28]. The specimens of this study were collected in Villefranche-sur-Mer (France) and most likely belonged to lineage L1, since all 234 individuals collected from eight locations at the French Mediterranean coast belonged to L1 (unpublished data). Here, we describe the reproduction of lineage L3 that occurred most likely at the beginning of May rather than at the end of May – early June, as previously reported [30]. The juveniles are then brooded for several weeks, unless a particular stress makes the mother expel the juveniles [30]. We did not observe fertilization events, but hypothesize that internal fertilization may occur by intake of sperm into the female bursae, as suggested by Byrne [50] and Byrne et al. [51] for Ophionereis olivacea (H.L. Clark, 1900) and Ophiopeza spinosa (Ljungman, 1867). The development of juveniles was apparently direct, but we cannot rule out the presence of an extremely transient brooded larval stage, which was not observed. Indeed, brooded vitellaria larvae were observed in ophiodermatid brittle stars [51].
Our results suggest that, in addition to their divergent reproductive strategies, L1 and L3 may display pre-zygotic isolation as fertilization in L3 individuals occurred before the beginning of May, whereas L1 did not spawn, even when stimulated, until the beginning of June. Although reproduction and gonad development should be followed over a year for both lineages, and at different geographic locations to confirm our results, the genetic data support reproductive isolation of L1 and L3 in Greece. Indeed, they displayed very divergent mitochondrial lineages, different haplotype diversities and different patterns of polymorphism at nuclear marker i51. The latter was polymorphic for the L1 broadcast spawning lineage, displaying 12 alleles of which 11 were private alleles, whereas i51 was monomorphic for the L3 brooding lineage, indicating a loss of genetic diversity in the brooding lineage. In addition, Boissin et al. [31] showed that two additional sympatric populations of L1 and L3 in Greece were differentiated for the nuclear marker ITS1.
The morphological as well as genetic and reproductive time differences between the lineages L1 and L3 of O. longicauda establish that those lineages are different biological species. To ensure that there is no possibility of hybridization between L1 and L3 because of a second reproductive period at other times of the year, a year-long study of the gonad condition should be performed. Fenaux [29], however, observed that there was only one event of reproduction in the year, at least in the lineage L1. In addition, the genetic data allow us to safely state that there is no longer gene flow between L1 and L3 in Crete. Further study is required to test whether the reproductive times can change and overlap in other places of the Mediterranean, due to differing environmental conditions.
In their study, Boissin et al. [31] described the occurrence of six divergent mitochondrial lineages in the O. longicauda species complex. To fully resolve the speciation events in O. longicauda, it remains unknown whether the lineages L2, L4, L5 and L6 belong to a single broadcasting species including L1, or a single brooding species including L3, or if each additional lineage corresponds to another biological species. Only after that, the taxonomy should be revised by properly describing one or several new biological species. However, it is quite clear after these and the previous studies that “O. longicauda” includes at least two species. We suggest treating the groups of broadcasting and brooding lineages as a separate morphospecies or a morphologically similar species complex.
Acknowledgments
We are very grateful to Christos Arvanitidis, without whom this study would not have been possible. We would like to thank him for his availability and his constant willingness to help. Many thanks to Thanos Dailianis, Elena Sarropoulou, and Magdalini Christodoulou for help during the sampling sessions. Thank you also to Christian Marschal and Chantal Bezac for help with the light and scanning electron microscopy, and thank you to Holly Roussel Perret-Gentil for language editing. Finally, we would like to thank one anonymous reviewer for useful comments to improve the manuscript.


