1 Introduction
Environmental stress causes non-desirable effects on plants’ growth and productivity, especially drought and salinity [1]. Synthesizing and accumulating compatible osmolytes in plants, such as proline and glycine betaine, facilitate coping with this condition [2,3].
Amino acid proline is an α-amino acid, and is not an essential amino acid, which means that living organisms can synthesize it. It is unique among amino acids, because it contains a secondary amino group. In addition to its role in protein forming, proline is one of the most widely distributed compatible solutes that accumulate in plants and bacteria during unfavorable environmental conditions [4,5]. The role of proline in the endurance of the environmental stress is still a matter of intensive research [6–8].
In plants, the synthesis of proline depends on two different precursors, glutamate and ornithine, through two different cycles [9,10]. In the first cycle, proline is produced via two reduction reactions of glutamate in which two enzymes catalyze these reactions, e.g., Δ-pyrroline-5-carboxylate synthase (P5CS) and pyrroline-5-carboxylate reductase (P5CR). P5CS is an enzyme activating glutamate through the phosphorylation process. This enzyme also reduces the product to form glutamate semi-aldehyde (GSA) [11]. In the second cycle of proline synthesis, ornithine turns to form pyrroline-5-carboxylate with the catalysis of orn-Δ-aminotransferase (OAT). This enzyme exists in mitochondria [7]. However, when plants experience adverse environmental conditions, proline is synthesized mainly through the first cycle. This has been demonstrated through analyses of the expression of P5CS and P5CR in Arabidopsis thaliana and moth bean plants [11–13].
Calotropis procera (C. procera) is a drought-tolerant wild plant. It belongs to the Asclepiadaceae family and is characterized as a sustainable evergreen toxic shrub. Seed spreads mainly by wind and can be transmitted by animals as well. Therefore, this plant is seen along roadsides, and the edges of lakes and native pastures, while scattered in desert areas [14,15]. C. procera is native to west and east Africa, and south Asia, while naturalized in Australia, Central and South America, and the Caribbean islands [15–17]. It provides an excellent source of genes for drought and salt tolerance. In previous work, we found that proline is increased in this plant when irrigated [18]. This finding is contrary to the conclusions of most researchers [2,3]. We suggest that this plant might need proline in another pathway under temporary irrigation. However, the biological significance of P5CS in C. procera has not been described. Increasing information about plant genomes in conjunction with bioinformatics tools and databases has led to the availability of new insights into the study of different genes that may be keys to stress responses in plant [19,20].
In this study, we uncovered and characterized one P5CS-like gene in this medicinal plant from the de novo assembled transcriptome contigs of a high-throughput sequencing dataset. We also compared the sequence as well as the three-dimensional (3D) structure of the obtained P5CS-like protein with those of other plant species.
2 Materials and methods
2.1 Sample collection and isolation of total RNA
Three leaf discs of C. procera were collected from Jeddah region (KSA, latitude 21°26′6.00, longitude 39°28′3.00 in September 2012 (with temperature of 37 °C, and air humidity of 70–75%). The samples were frozen in liquid nitrogen (50 mg tissue each) and total RNA extraction was performed using RNeasy Plant Mini Kit (Qiagen, cat. No. 74903). To remove DNA contaminants, 3 μL of 10 mg/mL RNase A, DNase and protease-free Thermo Scientific cat No. EN0531) were added to the RNA samples, and the tube was incubated at 30 °C for 15 min. The RNA concentration in different samples was estimated by measuring the optical density at 260 nm according to the equation: RNA concentration (μg/mL) = OD260 × 40 × dilution factor. RNA samples were sent to Beijing Genomics Institute (BGI), Shenzhen, China, for deep sequencing, and dataset were provided for analysis.
2.2 NGS sequence
Whole-RNA-seq, paired-end short-sequence reads of C. procera were generated using the Illumina Genome AnalyserIIx (GAIIx) according to the manufacturer's instructions (Illumina, San Diego, CA).
2.3 Sequence filtering and bioinformatics analysis
The raw sequencing data were obtained using the Illumina python pipeline v. 1.3. For the obtained libraries, only high-quality reads (quality > 20) were retained. Then, a de novo assembly of the obtained short (paired-end) read dataset was performed using assembler trinityrnaseq_r20131110 [21] followed by the creation of putative unique transcripts (PUTs) with a combination of different k-mer lengths and expected coverage.
Twenty P5CS sequences (Table 1) belonging to other plant species were obtained from GenBank and used as a reference for blasting (http://www.ncbi.nlm.nih.gov/BLAST) our obtained library (the yielded EST assemblies from Velvet program) to identify contigs with CpP5CS-like sequence.
Accession numbers, description of the gene and organism whose P5CS-like gene was isolated.
| Accession no. | Description | Organism |
| ABO70348.1 | Pyrroline-5-carboxylate synthetase | Apocynum venetum |
| XP_006346827.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-like | Solanum tuberosum |
| AEN04068.1 | Δ1-pyrroline-5-carboxylate synthetase | Solanum torvum |
| XP_006355262.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-like | Solanum tuberosum |
| ADL61840.1 | Δ1-pyrroline-5-carboxylate synthetase | Nicotiana tabacum |
| NP_001233907.1 | Δ1-pyrroline-5-carboxylate synthase | Solanum lycopersicum |
| AAC14481.1 | Pyrroline-5-carboxylate synthetase | Actinidia deliciosa |
| CBI31612.3 | Unnamed protein product | Vitis vinifera |
| XP_004240687.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-like | Solanum lycopersicum |
| ABG74923.1 | Pyrroline-5-carboxylate synthetase | Aegiceras corniculatum |
| EOY07413.1 | Pyrroline-5-carboxylate synthetase isoform 1 | Theobroma cacao |
| XP_004138450.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-like | Cucumis sativus |
| AHF58596.1 | Δ1-pyrroline-5-carboxylate synthetase 2 | Chrysanthemum lavandulifolium |
| ACI62865.1 | Δ1-pyrroline-5-carboxylate synthetase | Gossypium arboretum |
| AEO27874.1 | Pyrroline-5-carboxylate synthetase | Cucumis melo |
| NP_001268134.1 | Pyrroline-5-carboxylate synthetase | Vitis vinifera |
| XP_003519362.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-like | Glycine max |
| XP_003544177.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-like | Glycine max |
| AGW51410.1 | Pyrroline-5-carboxylate synthetase | Salicornia bigelovii |
| ABZ79407.2 | Pyrroline-5-carboxylate synthetase | Gossypium arboreum |
Assemblies were mapped to Apocynum venetum accession number EF160132 using SAOP [22]. The number of reads aligned was 6577, with an average coverage of 327.33 and the length of consensus sequence, including C. procera P5CS-like (CpP5CS-like) gene, equals 2154 nt (Fig. 1).

(Color online). ORF analysis of the obtained CpP5CS-like sequence.
2.4 Determination of phylogenetic relationships
The maximum-likelihood method [23] was used to build a dendrogram and CLC Genomics Workbench was used to allow doing bootstrap analysis. A bootstrap value is attached to each branch to indicate the confidence level in this branch.
2.5 The 3D homology modeling
Homology modeling was carried out using Swiss-Model, a protein-modeling server, accessible via the EXPASY (http://www.expasy.org/). Superimposition of CpP5CS-like amino acid sequence model on those of other P5CS proteins was constructed using RasMol (http://www.umass.edu/microbio/rasmol/), and Deep-View programs (http://spdbv.vital-it.ch/). The functional domains were identified from the NCBI's conserved domain database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), which uses 3D structure information to explicitly define domain boundaries and provide insights into sequence/structure/function relationships, as well as domain models imported from a number of external source databases (Pfam, SMART, COG, PRK, TIGRFAM).
2.6 Structure alignment
The protein model was applied to the pairwise comparison of protein structures using the structure domains of DaliLite program (server at EBI, https://www.ebi.ac.uk/Tools/dalilite/) [24]. Root mean square deviation (RMSD), which measures the average distance between the backbones of superimposed proteins, was measured according to the following formula:
3 Results and discussion
To allocate protein domains, the protein sequence obtained from ORF analysis with a length of 717 was analyzed against the CDD database (conserved domain database, http://www.ncbi.nlm.nih.gov/cdd) to detect protein domains. Domain analysis indicated the presence of two protein domains (AAK_P5CS_ProBA conserved domain, database accession number CD04256 and ALDH conserved domain, database accession number CD07079).
3.1 BLAST analysis
To identify sequence similarities with homologous proteins from other organisms, PHI-BLAST and DELTA-BLAST tools were performed to the obtained C. procera P5CS protein (http://blast.ncbi.nlm.nih.gov/). The explanation of the score and sequence similarity from specialized BLAST searching eventually led to the identification of putative or homologous protein sequences. Our results for the most closely related protein to C. procera P5CS protein indicated that the PREDICTED pyrroline-5-carboxylate synthase-like of Apocynum venetum has the lowest e-value (0.0) and a high identity percent. These results indicate that C. procera P5CS has a same function.
3.2 Multi-sequence alignment (MSA) and phylogenetic analysis
The best BLAST search hits were used to perform multi-sequence alignment (Table 2). This resulted in 21 P5CS protein sequences from 15 different species, including C. Procera. A multiple sequence alignment of the 21 sequences was obtained by a gap-opening penalty of 10 and a gap extension penalty of one (Fig. 2). Twenty sequences with the obtained C. procera P5CS protein were used to perform pair wise alignment (Table 3). The results also show that the closest sequence to the obtained C. procera P5CS protein is Apocynum venetum PREDICTED: the PREDICTED pyrroline-5-carboxylate synthase with accession number ABO70348.1. These results support the obtained BLAST results. MSA results were used to perform a phylogenetic tree for the 20 proteins and results (Fig. 3) were similar to those of previous analyses.
Accession number for each protein, description, organism name and the calculated e-value of homologous proteins to C. procera P5CS amino acids sequence identified using specialized BLAST search programs.
| Accession | Description | T.S. | Q.C. (%) | e-value | Max. ident (%) |
| ABO70348.1 | Pyrroline-5-carboxylate synthetase [Apocynumvenetum] | 1316 | 99 | 0 | 94 |
| XP_006346827.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik…. | 1233 | 99 | 0 | 84 |
| AEN04068.1 | Δ1-pyrroline-5-carboxylate synthetase [Solanumtorvum] | 1229 | 99 | 0 | 85 |
| XP_006355262.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik…. | 1228 | 99 | 0 | 85 |
| ADL61840.1 | Δ1-pyrroline-5-carboxylate synthetase [Nicotianataba…. | 1219 | 99 | 0 | 85 |
| NP_001233907.1 | Δ1-pyrroline-5-carboxylate synthase [Solanumlycop…. | 1216 | 99 | 0 | 84 |
| O04015.1 | Δ1-pyrroline-5-carboxylate synthase [Actinidiadeliciosa] | 1214 | 99 | 0 | 86 |
| CBI31612.3 | Unnamed protein product [Vitisvinifera] | 1211 | 99 | 0 | 82 |
| XP_004240687.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik…. | 1204 | 99 | 0 | 84 |
| EOY07413.1 | Pyrroline-5-carboxylate synthetase isoform 1 [Theobroma…. | 1187 | 99 | 0 | 81 |
| XP_004138450.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik… | 1182 | 99 | 0 | 82 |
| AHF58596.1 | Δ1-pyrroline-5-carboxylate synthetase 2 [Chrysa…. | 1175 | 99 | 0 | 82 |
| ACI62865.1 | Δ1-pyrroline-5-carboxylate synthetase [Gossypium…. | 1169 | 98 | 0 | 83 |
| AEO27874.1 | Pyrroline-5-carboxylate synthetase [Cucumismelo] | 1168 | 99 | 0 | 82 |
| NP_001268134.1 | pyrroline-5-carboxylate synthetase [Vitisvinifera] | 1166 | 99 | 0 | 81 |
| XP_003519362.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik… | 1165 | 98 | 0 | 81 |
| XP_003544177.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik…. | 1165 | 99 | 0 | 81 |
| AGW51410.1 | Pyrroline-5-carboxylate synthetase [Salicorniabigelovii] | 1163 | 99 | 0 | 79 |
| ABZ79407.2 | Pyrroline-5-carboxylate synthetase [Gossypiumarboreum] | 1290 | 99 | 0 | 83 |
| XP_004961886.1 | PREDICTED: Δ1-pyrroline-5-carboxylate synthase-lik…. | 1161 | 99 | 0 | 78 |

(Color online). Multiple sequence alignment of the 20 different P5CS protein sequences with the obtained C. procera P5CS protein sequence.
Pairwise alignment between each hit P5CS sequence as compared to the obtained sequence of Calotropis P5CS amino acids sequence.
| Accession number | Gaps | Alignment length | Differences | Query cover (%) | Identity % |
| ABO70348.1 | 0 | 714 | 3 | 99 | 94.12 |
| XP_006346827.1 | 0 | 714 | 3 | 99 | 84.17 |
| AEN04068.1 | 0 | 715 | 2 | 99 | 85.17 |
| XP_006355262.1 | 0 | 712 | 5 | 99 | 85.11 |
| ADL61840.1 | 0 | 712 | 5 | 99 | 84.69 |
| NP_001233907.1 | 0 | 712 | 5 | 99 | 84.27 |
| O04015.1 | 0 | 716 | 1 | 99 | 85.89 |
| CBI31612.3 | 0 | 715 | 2 | 99 | 82.24 |
| XP_004240687.1 | 0 | 714 | 3 | 99 | 84.45 |
| EOY07413.1 | 0 | 710 | 7 | 99 | 84.37 |
| XP_004138450.1 | 0 | 714 | 3 | 99 | 81.37 |
| AHF58596.1 | 0 | 714 | 3 | 99 | 82.35 |
| ACI62865.1 | 0 | 711 | 6 | 98 | 81.86 |
| AEO27874.1 | 0 | 709 | 8 | 99 | 83.07 |
| NP_001268134.1 | 0 | 710 | 7 | 99 | 81.69 |
| XP_003519362.1 | 0 | 711 | 6 | 98 | 80.59 |
| XP_003544177.1 | 0 | 709 | 8 | 99 | 81.38 |
| AGW51410.1 | 0 | 711 | 6 | 99 | 81.43 |
| ABZ79407.2 | 0 | 712 | 5 | 99 | 78.51 |
| XP_004961886.1 | 0 | 709 | 8 | 99 | 82.65 |
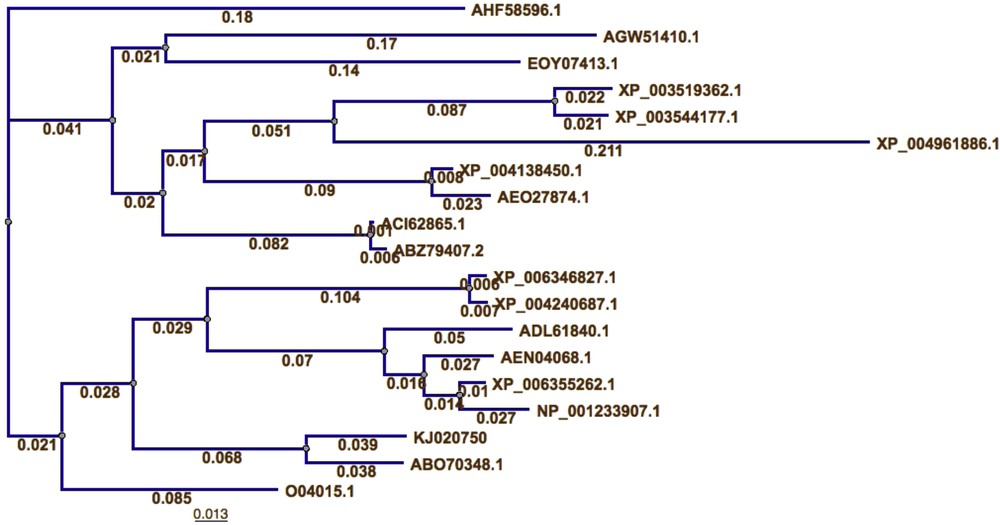
(Color online). Phylogenetic analysis of 20 P5CS proteins and C. procera P5CS deduced amino acids sequence (accession No. KJ020750).
3.3 3D structure modeling
P5CS signaling efficiency and specificity can be achieved through human pyrroline-5-carboxylate synthetase (http://www.ebi.ac.uk/pdbe-srv/view/entry/2h5g/summary).
Based on structural alignment, a theoretical 3D model for C. procera P5CS protein was created, corresponding to residues 1–717 of the primary structure (Fig. 4). The predicted model was created using the Swiss-Model protein-modeling server.
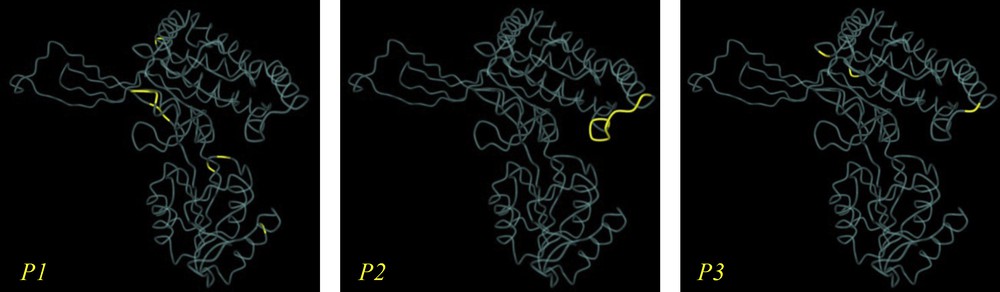
(Color online). Predicted 3D model of deduced amino acids sequence of C. procera P5CS (accession No. KJ020750), including all important functional domains and motifs. P1, putative nucleotide binding site, based on the similarity to Campylobacter jejune glutamate 5-kinase. P2, putative phosphate binding site, based on the similarity to the region identified in tomato glutamate 5-kinase. P3, putative allosteric binding site, based on similarity to mutational studies in tomato glutamate 5-kinase.
The overall model dimensions are 122.022 Å × 137.402 Å × 72.057 Å.
3.4 Structure alignment
We applied DaliLite on the 3D structures of nine proteins, which were created based on structural alignment using Swiss-Model.
2h5g, human pyrroline-5-carboxylate synthetase, is the closest homologous protein sequence with available 3D structure to the obtained C. procera P5CS; however, 2h5g is a human P5CS also known as P5CS_HUMAN.
To prove the accuracy of our theoretical 3D model, we used DaliLite to compute optimal and suboptimal structural alignments between 2h5g 3D structure and the theoretical 3D model of C. procera P5CS protein. The resulting superimposed figure is shown in Fig. 5 with a Z-score of 58.7, number of equivalent residues of 406 and RMSD of 0.7.
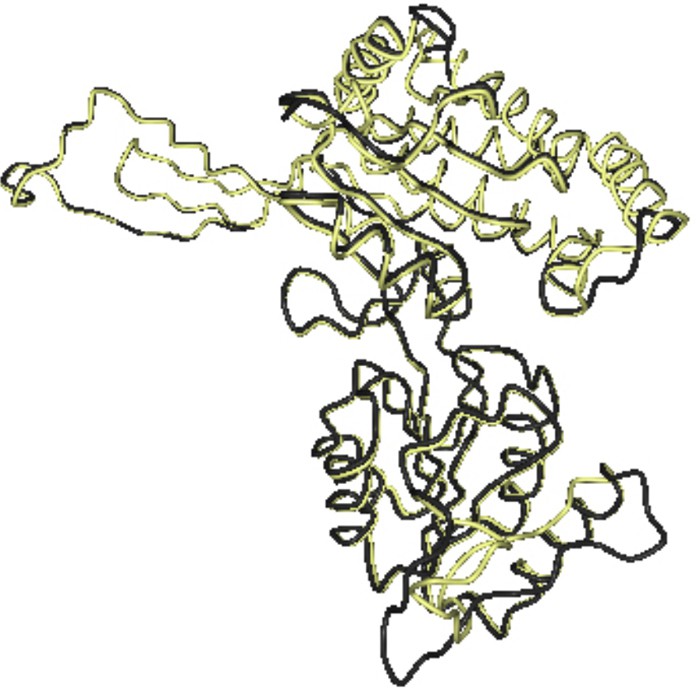
(Color online). 3D model of C. procera: P5CS deduced amino acids (yellow) have almost the same coordinates as 2h5g (gray).http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
4 Discussion
The obtained C. procera sequence showed features Δ-pyrroline-5-carboxylate synthase (P5CS). P5CS protein domain whose features determine its function has been evolutionarily conserved in numerous eukaryotic organisms and partially in prokaryotic. Δ-pyrroline-5-carboxylate synthases are known to provide plants with the ability to overexpress in response to environmental stresses, such as nutrient starvation, drought and high salinity. These conditions are normal for a desert wild plant, like C. procera.
4.1 Conserved domain analysis
Domain analysis indicated the presence of two protein domains. First, AAK_P5CS conserved domain database accession number CD04256, and pfam accession number PF00696.23. And the second is ALDH_F1819_ProA-GPR conserved domain database accession number CD07079, and pfam database accession number PF00171.17 (Fig. 6). To assign the identified Δ-pyrroline-5-carboxylate synthase (P5CS) and its appropriate protein subfamilies, several further analyses were conducted. First, we made a BLAST search against GenBank protein database. The interpretation of the score, query coverage, e-value and sequence identity, led to the identification of putative homologous protein sequences. The results showed that the most closely related gene was pyrroline-5-carboxylate synthase-like of Apocynum venetum, which has the lowest e-value (00) and high percent identity. These results indicate that the speculated C. procera P5CS protein is almost typical of the isolated P5CS from another organism.
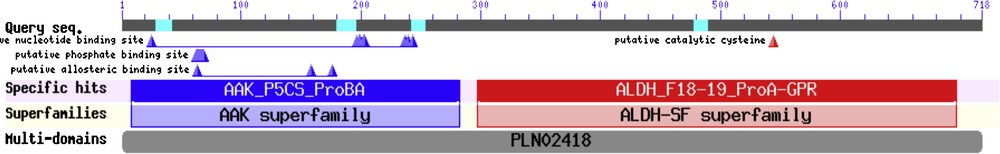
(Color online). Protein domains of the deduced amino acid sequence of the obtained P5CS protein.
The best BLAST search hits were used to perform multi-sequence alignment and 20 sequences resulted. The alignment of the 21 sequences (Table 2 and Fig. 2) showed that the closest sequence to the obtained C. procera P5CS amino acids sequence is Apocynum venetum pyrroline-5-carboxylate synthetase (accession No. ABO70348.1). MSA results were used to draw a phylogenetic tree for the 21 P5CS proteins, and the results (Fig. 3) were similar to those of previous analyses. Multi-sequence alignment also proved that all important functional domains and motifs belonging to P5CS are located within C. procera P5CS deduced amino acids sequence (Fig. 4, P1, P2, P3).
P5CS includes two functional conserved domains. AAK superfamily [cl00452], which is the amino acid kinases (AAK) superfamily catalytic domain; glutamate-5-kinase (G5 K) domain of the bifunctional Δ1-pyrroline-5-carboxylate synthetase (P5CS), composed of an N-terminal G5 K (ProB) and a C-terminal glutamyl 5-phosphate reductase (G5PR, ProA), the first and second enzyme catalyzing proline. G5 K transfers the terminal phosphoryl group of ATP to the gamma-carboxyl group of glutamate, and is subject to feedback allosteric inhibition by proline or ornithine. In plants, proline plays an important role as an osmo-protectant [25–27].
4.1.1 Putative 1 (P1)
It is an autative nucleotide binding site [chemical binding site], based on the similarity to Campylobacter jejune glutamate 5-kinase.
AA no. 18, 209-211, 214-215, 249, 251, 274
4.1.2 Putative 2 (P2)
It is a putative phosphate binding site [ion binding site], based on the similarity to the region identified in tomato glutamate 5-kinase.
AA Nos. 56–63
4.1.3 Putative 3 (P3)
It is a putative allosteric binding site, based on the similarity to mutational studies in tomato glutamate 5-kinase.
AA Nos. 57, 136, 188
The second domain is gamma-glutamyl phosphate reductase (GPR), aldehyde dehydrogenase families 18 and 19, which is a part of a hierarchy of related CD model the superfamily NAD(P)+-dependent aldehyde dehydrogenase superfamily, CDD No. accession cl11961.
Gamma-glutamyl phosphate reductase (GPR) is an l-proline biosynthetic pathway (PBP) enzyme that catalyzes the NADPH dependent reduction of l-gamma-glutamyl 5-phosphate into l-glutamate 5-semi-aldehyde and phosphate.
The glutamate route of the PBP involves two enzymatic steps catalyzed by gamma-glutamyl kinase (GK, EC 2.7.2.11) and GPR (EC 1.2.1.41). These enzymes are fused into the bifunctional enzyme, ProA or Δ1-pyrroline-5-carboxylate synthetase (P5CS) in plants and animals, whereas they are separate enzymes in bacteria and yeast. In humans, the P5CS (ALDH18A1), an inner mitochondrial membrane enzyme, is essential to the de novo synthesis of the amino acids proline and arginine. Tomato (Lycopersicon esculentum) has both the prokaryotic-like polycistronic operons encoding GK and GPR (PRO1, ALDH19) and the full-length, bifunctional P5CS (PRO2, ALDH18B1).
4.1.4 Putative 1
Putative catalytic cysteine [active site] is a conserved cysteine that aligns with the catalytic cysteine of the ALDH superfamily.
Moreover, Fig. 5 shows that 3D model of C. procera P5CS amino acids (yellow) has almost the same coordinates of 2h5g (gray).
These results support our finding that the obtained C. procera protein sequences belong to P5CS and possess the same functions regarding its functional domains and that the motifs belong to P5CS. Also, the results prove the accuracy of our theoretical 3D modeling for the obtained C. procera P5CS deduced from the sequence of the amino acids. Further study to detect the regulation of this gene under abiotic stress conditions is underway.
As a conclusion, the present study provides two results. First, our study reports an important gene involved in environmental stress in the arid land plant Calotropis procera and it is first record reporting the presence of P5CS. Second, our study provides sufficient information for future manipulations of P5CS gene expression, which may explain the molecular bases of the increase in proline accumulation in this plant under watering conditions, in contrast with its known role in other plants, where it usually accumulates in plants under drought conditions. Moreover, our study proves that gene structure and function are not related to this observation. Further studies may lead to a better understanding of this phenomenon in Calotropis procera.
Acknowledgements
The authors gratefully acknowledge the financial support from King Abdulaziz University (KAU), and its Vice-President for Educational Affairs, Prof. Dr. Abdulrahman O. Alyoubi. Also, Prof. Dr. Gamal Saber and Prof. Dr. Ahmed Bahieldin (section of Genomics and Biotechnology) are acknowledged for support. The authors want to pay tribute to the skills of their dear colleague Dr. Ahmed Shokry who taught them what bioinformatics means.


