1 Introduction
CMYA1 (cardiomyopathy-associated protein1), also named Xin, which means ‘heart’ in Chinese, is specifically expressed in striated muscles. CMYA1 was first identified in the intercalated disk of chicken cardiomyocytes during a screen of mRNA from chicken embryos using differential display PCR [1–4]. CMYA1 is a downstream target of the transcription factors Nkx2.5 and Mef2 in the BMP-Nkx2.5-Mef2 C pathway and has been identified as a muscle-damage specific marker because of the important role it plays in myofibril assembly and muscle recovery [1,5–7]. CMYA1 localizes to the adherens junctions in cardiac and skeletal muscle, and mouse CMYA1 co-localizes with N-cadherin and β-catenin throughout embryogenesis and into adulthood and is responsible for crosslinking actin filaments into a loosely packed meshwork [8,9]. The CMYA1 protein contains an amino-terminal nuclear localization signal (NLS), a DNA-binding domain (DBD), several proline-rich regions that interact with both EVH1 domains and SH3 domains of nebulin, and a varying number of novel 16 amino acid (aa) repeats (here in named Xin-repeats). Xin-repeats is a highly evolutionarily conserved region in mammalian CMYA1 proteins and the consensus sequence is GDV(K/Q/R/S)XX(R/K/T)WLFET(Q/R/K/T)PLD [10–13].
Research on CMYA1 interacting proteins is necessary to characterize its functions. Mammalian CMYA1 has been shown to bind certain components of the cardiac Z-disc, such as a-actinin, filaminC, conneXin43, aciculin, as well as vasodilator-stimulated phosphoprotein (VASP) and myopodin [9,14,15]. The Xin-repeats of CMYA1 is a significant domain that occupies more than half of the protein length and has been shown to play an important role in protein interactions [10,15]. The Xin-repeats can interact with α-actin and β-catenin within the intercalated discs, where they function to stabilize the actin-based cytoskeleton [15]. Interestingly, using different truncated mouse Xin-repeats, it was found that the ability of the Xin-repeats to bind actin filaments is correlated with the number of serial Xin-repeats, with at least three repeats being necessary for actin binding and six for binding to actin filaments [11,15]. The binding force was multiplied as the numbers of Xin-repeats increased.
To date, there has been notably little study of bovine CMYA1. As mentioned above, CMYA1 functions during muscle development and the Xin-repeats plays a significant role in the function of CMYA1. Thus, to identify the function of CMYA1/Xin-repeats in bovine muscle development, we used CMYA1/Xin-repeats as targets in a yeast two-hybrid assay to find more proteins that interacted with CMYA1/Xin repeats in bovine muscle. Previously, we obtained the full-length cDNA sequence of bovine CMYA1 and submitted it to Genbank with the accession number HQ111436.1. Bovine CMYA1 maps to chromosome 22 and contains two exons, of which the second exon contains the complete coding sequence of the CMYA1 protein.
In this study, we aimed to identify novel proteins that interact with CMYA1/Xin-repeats to help identify certain of the important regulatory networks involved in muscle development. The yeast two-hybrid (Y2H) system is one of the most useful, cost-effective and widely used methods for high-through put protein–protein interaction (PPI) studies in vivo [7,16]. Presently, there is no bovine muscle cDNA library available that can be used for detecting interacting proteins. In this study, we constructed a high-quality cDNA library of bovine foreleg muscle and two bait vectors (CMYA1 and Xin-repeats) for a GAL4-based Y2H system. Through two Y2H screens, 27 candidate interacting proteins of CMYA1 and Xin-repeats have been identified from the bovine muscle cDNA library. Three of the 27 candidate proteins (RPL35A, RPL21 and EIF3G) interacted with both CMYA1 and Xin-repeats, and the interaction was further confirmed using their full-length coding DNA sequences (CDS) as prey in a one-to-one Y2H. Thus, we have validated three putative interacting proteins of CMYA1/Xin repeats that had not been previously reported. These results suggest that CMYA1/Xin repeats may be required in the protein synthesis and cells growth pathways and further demonstrate that the Xin-repeat is a key domain in CMYA1 function. This study provides a new insight into the mechanisms of bovine muscle development, and this information could be used to help improve bovine breeding.
2 Materials and methods
2.1 Ethics approval statement
All animal sample procedures were approved and performed in accordance with institutional guidelines.
2.2 Animal and tissue samples
Bovine tissue specimens, weighing 200–300 mg per sample, were obtained in triplicate from the foreleg muscle of a single adult male Simmental. The samples were immediately stored in liquid nitrogen and were moved into the freezer at –80 °C to be used for the experiment. All animal sample procedures were approved and performed in accordance with institutional guidelines.
2.3 Total RNA isolation and reverse-transcription into cDNA
High-quality total RNA was extracted from bovine foreleg muscle using the TRIzol Reagent (Invitrogen, USA). Briefly, approximately 1 mg of total RNA from muscle tissue was used to isolate polyA mRNA. The first-strand cDNA was synthesized using the All-in-one™ First-Strand cDNA Synthesis Kit (GeneCopoeia) according to the manufacturer's protocol. The ds-cDNA (double stranded-cDNA) was then divided into two parts, one was used to prepare clones in the pGADT7-Rec vector, and the other was used for PCR-amplification of full-length CMYA1 and the Xin-repeat fragment of CMYA1.
2.4 Construction of bovine muscle cDNA library
To construct the bovine muscle cDNA GAL4-AD yeast two-hybrid mating library, the yeast Y187 strain (Clontech) was co-transformed with the purified ds-cDNAs and the linearized pGADT7-Rec vector using the Make Your Own “Mate & Plate™” Library System technology (Cat.No.630490). Selection was performed using SD/-Leu agar medium. To identify the cDNA fragment length, a certain number of colonies were picked randomly and PCR-amplified with the common primers (AD-F CGCGTTTGGAATCACTACAGGG and AD-R GGAGACTTGACCAAACCTCTGGCG) (Oligo Synthesis was in Genewiz). The number of clones screened was calculated by counting the colonies from the DDO plates (dropout medium that lacks both leucine and tryptophan). No. of screened clones = No. of cfu/ml of diploids × resuspension volume (mL) (standard ≥ 1 million cfu). Mating Efficiency% = No. of CFU/mL of limiting partner × 100 (5% ≥ standard ≥ 2%). The resultant transformants were pooled and frozen stored at –80 °C. After thawing, the library titer was calculated. Library titer = No. of CFU/mL of diploids (standard ≥ 1 × 107 CFU/mL).
2.5 Construction of CMYA1/Xin repeats bait vectors
To construct the yeast expression plasmids, NCBI BLAST was used to identify the conserved Xin-repeat region (144–2612 bp). Using the bovine muscle cDNA as a template, the full-length CMYA1 CDS (144–5606 bp) were PCR-amplified and directionally cloned into the pGBKT7 vector (Clontech) usingthe EcoRI and SalI restriction sites. The Xin-repeat region (144–2612 bp) was cloned using the EcoRI and BamHI restriction sites (Fig. 1A, Table 2). The plasmids pGBKT7-CMYA1 and pGBKT7-Xin repeats were transformed into DH5α competent cells for DNA preservation and proliferation. The resulting plasmids were transformed into the Y2HGold strain following the Clontech Yeastmaker™ Yeast Transformation System 2 User Manual (Cat#630439). To test the autoactivation and the toxicity of the bait proteins, yeast transformed with 100 ng of the bait vectors for CMYA1/Xin repeats were grown on SD/-Trp (dropout medium lacking tryptophan), or SD/-Trp/X (SD/-Trp supplemented with 40 μg/mL X-α-Gal), or SD/-Trp/X/A (SD/-Trp supplemented with 40 μg/mL X-α-Gal and 125 ng/mL aureobasidin A) at 30 °C for 3–5 days. The growth of white colonies on SD/-Trp and SD/-Trp/X mediums and no colony growth on SD/-Trp/X/A plates could indicate no autoactivation. White colonies that grow to a similar size, and at the same speed, as the pGBKT7 vector could indicate that there is no toxicity. Only bait vectors that show no autoactivation activity or toxicity could be used for the Y2H screen.
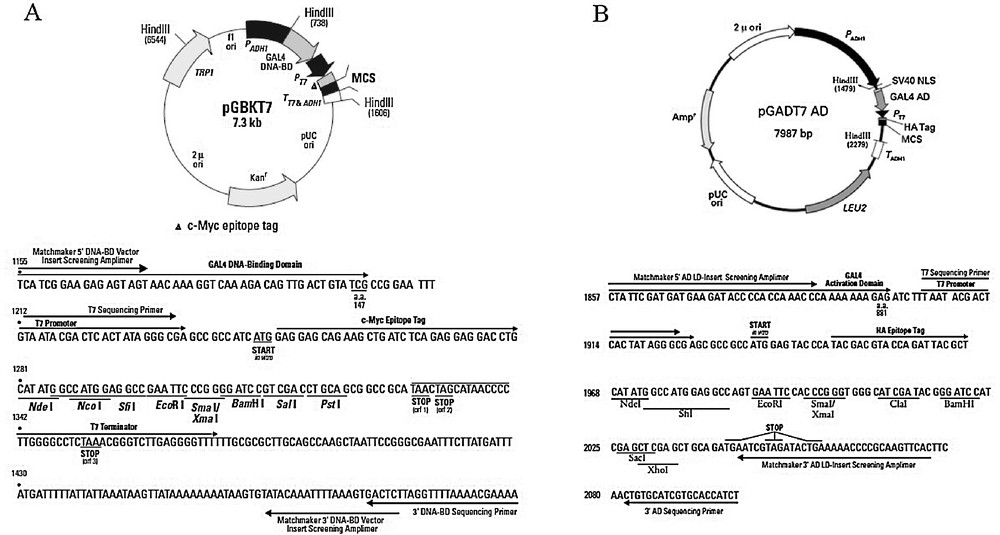
Plasmid diagram. (A) Bait plasmid. The Xin repeats fregment was cloned into the pGBKT7 vector using the EcoRI and BamHI restriction enzymes, and EcoRI and SalI were used for the cloning of CMYA1 in pGBKT7. (B) Prey plasmid. The full-length CDS of RPL35A was cloned into pGADT7 vector using Nde1 and BamH1. NdeI and EcoRI were used for the cloning of EIF3G, and NdeI and EcoRI were used for the cloning of RPL21.
Primer pairs designed for this experiment. Restriction sites are noted by italics.
| Primer name/Main target | Sequences (5’ → 3’) | Position of target | Size(bp) | |
| Xin repeat-EcoR I | Forward | CGGAATTCATGGCCAATGCCCAGACGCA | 144 | 2469 |
| Xin repeat-BamH I | Reverse | GCGGATCCCTCCTCCTTCCGAACATGG | 2612 | |
| CMYA1- EcoR I | Forward | CGGAATTCATGGCCAATGCCCAGACGCA | 144 | 5463 |
| CMYA1- Sal I | Reverse | GCGTCGACTGGTGGCAGATGGAGGGGCT | 5606 | |
| RPL35 A-Nde I | Forward | CGCATATGATGTCTGGAAGGCTGTGGTC | 63 | 333 |
| RPL35 A-BamH1 | Reverse | GCGGATCCTTAAATTCTCGAAGGGTACA | 395 | |
| EIF3G-Nde I | Forward | CGCATATGATGCCGACCGGAGACTTTGA | 49 | 963 |
| EIF3G-EcoR I | Reverse | CGGAATTCTTAATTGGTTGATGGCTTGG | 1011 | |
| RPL21-Nde I | Forward | CGCATATGATGACCAACACAAAGGGAAA | 286 | 483 |
| RPL21-EcoR I | Reverse | CGGAATTCTTTTTACACCCATCAGGCCA | 768 |
2.6 Identification of CMYA1/Xin repeats interacting proteins
In our study, we performed a yeast two-hybrid screen twice following the protocol of the Clontech Matchmaker™ Gold Yeast Two-Hybrid System (Cat.No. 630489). First, for the initial selection of positively interacting clones, the bait strains that contained the pGBKT7-CMYA1 or pGBKT7-Xin repeats vectors were co-cultured with the bovine muscle cDNA library in a high-concentration YPDA (2×) liquid medium ideal for yeast mating to the fusion cell, and selection was performed using DDO plates. The positive fusion colonies were identified because they could grow on DDO/X/A (dropout medium of lacking both leucine and tryptophan supplemented with X-α-Gal and aureobasidin A) and appeared blue because of reporter genes of Leu/Trp/X-a-Ga/Aba that were expressed when interactions occurred between bait and prey. To eliminate false positives, all positive colonies were transformed and plated onto a more restrictive dropout minimal media. All blue colonies were picked using sterile pipet tips and streaked onto QDO/X/A plates (dropout medium lacking adenine, histidine, leucine, and tryptophan, supplemented with X-α-Gal and Aureobasidin A).
2.7 Verification of candidate interacting proteins
To identify the preys that were genuine positives, the prey inserts were sequenced using primers for the T7 promoter and were BLASTed on NCBI. The sequencing results of the positive prey inserts were BLASTed against the bovine genome (Bos_taurus_UMD_3.1.1 reference Annotation Release 104) to identify the corresponding bovine genes to which the sequence reads were in-frame. The majority of interacting proteins identified were partial segments of the CDS. To verify binding between the positive prey proteins and the bait proteins, CMYA1 and Xin repeats, a one-to-one mating Y2H assay was performed following the small-scale transformation protocol. The completed CDs for three candidate prey proteins were cloned into the vector pGADT7 and transformed into the yeast strain Y187. The vectors were co-cultured with bait-CMYA1 or bait-Xin repeats in YPDA media and screened using DDO/X or QDO/X/A medium using pGBKT7–53 and pGADT7-T as positive controls and pGBKT7-Lam and pGADT7-T as negative controls. The transformants were plated on DDO/X or QDO/X/A medium for 3–5 days at 30 °C for selection. Construction of the pGADT7-prey vectors and transformation into the Y187 stain with the GAL-BD was performed using the same methods that were described before (Fig. 1B). Growth of blue colonies on both DDO/X and QDO/X/A/confirmed that the candidate prey protein interacted with the bait; in contrast, in the control group, only white colonies grew on DDO/X, and no colonies grew on QDO/X/A.
3 Results
3.1 Generation of bovine muscle cDNA library for Y2H system
Currently, to the best of our knowledge, there is no cDNA library of bovine muscle available to us for the detection of PPIs using the Y2H system. In this study, we have successfully generated a high-quality bovine muscle cDNA library for use in a GAL4 Y2H screen, which will be of great value to the study the regulatory networks that function during muscle development. The quality of the library was assessed by the following criteria: the number of clones screened, the sizes of the inserts, and recombination efficiency. The number of screened clones and recombination efficiency were calculated using gradient dilution and colony counting. In total, the volume of the cDNA library was 40 mL. The number of screened clones was 2.8 million, which satisfied the demand for more than 1 million clones (Fig. 2A, Table 1). To check the inserts size and to analyze the recombination efficiency, we performed PCR on approximately 100 random colonies from the library using pGADT7 specific primers (Table 2). Gel electrophoresis showed that the insert sizes of the muscle library ranged from 0.2 to 2.0 kb, and the average size was approximately 0.6 kb (Fig. 2C, Table 1). The mating efficiency was 3.27%, which satisfied the standard (2% ≥ standar ≥ 5%) (Fig. 5A, Table 1). The cDNA library titer was 3.5 × 107 CFU/mL (Fig. 2B, Table 1).
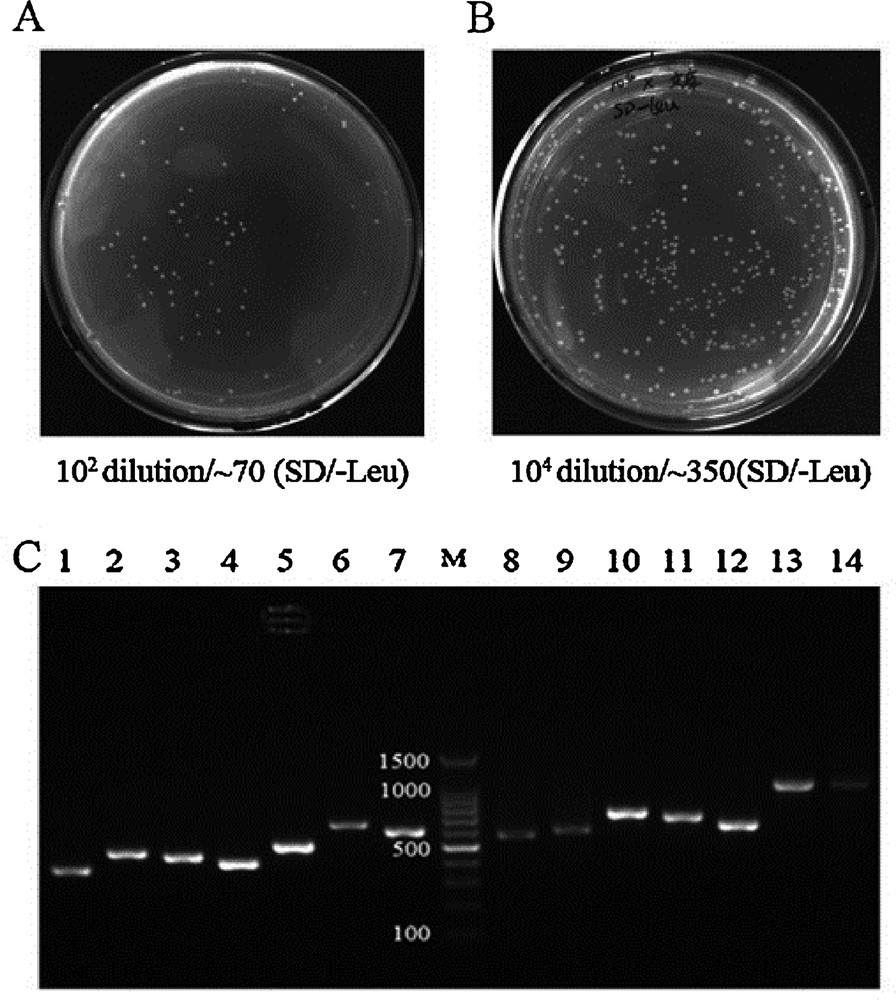
Quality of cDNA library. (A) The total volume of the library was 40 mL. Library cells (100 μL) were plated on SD/-Leu agar medium at a 102 dilution. More than 70 clones grew after 3–5 days at 30 °C. The total number of screened clones was 2.8 million (standard ≥ 1 million). (B) Thawing of the frozen library, and dilutionat different ratios. When 100 μL of the library was plated on SD/Leu, more than 350 colonies grew on a 104 dilution plate. The titer of the cDNA library was 3.5 × 107 (standard ≥ 1 × 107). (C) Representiveagarose gel analysis from 100 randomly selected colonies of the cDNA library. Masquer
Quality of cDNA library. (A) The total volume of the library was 40 mL. Library cells (100 μL) were plated on SD/-Leu agar medium at a 102 dilution. More than 70 clones grew after 3–5 days at 30 °C. The total number of screened clones ... Lire la suite
Characteristic of cDNA library was reflected by randomly picked 100 colonies.
| Bovine muscle cDNA library characteristic | |
| Bovine muscle cDNA library | |
| Number of screened clones (standard ≥ 1 million) (cfu) | 2.8 million |
| Titer of cDNA library (cfu/mL) (standard ≥ 1 × 107) | 3.5 × 107 |
| Mating Efficiency (2% ≥ standard ≥ 5%) | 3.27% |
| Minimum insert size (kb) | 0.2 |
| Maximum insert size (kb) | 2.0 |
| Average insert size (kb) | 0.6 |
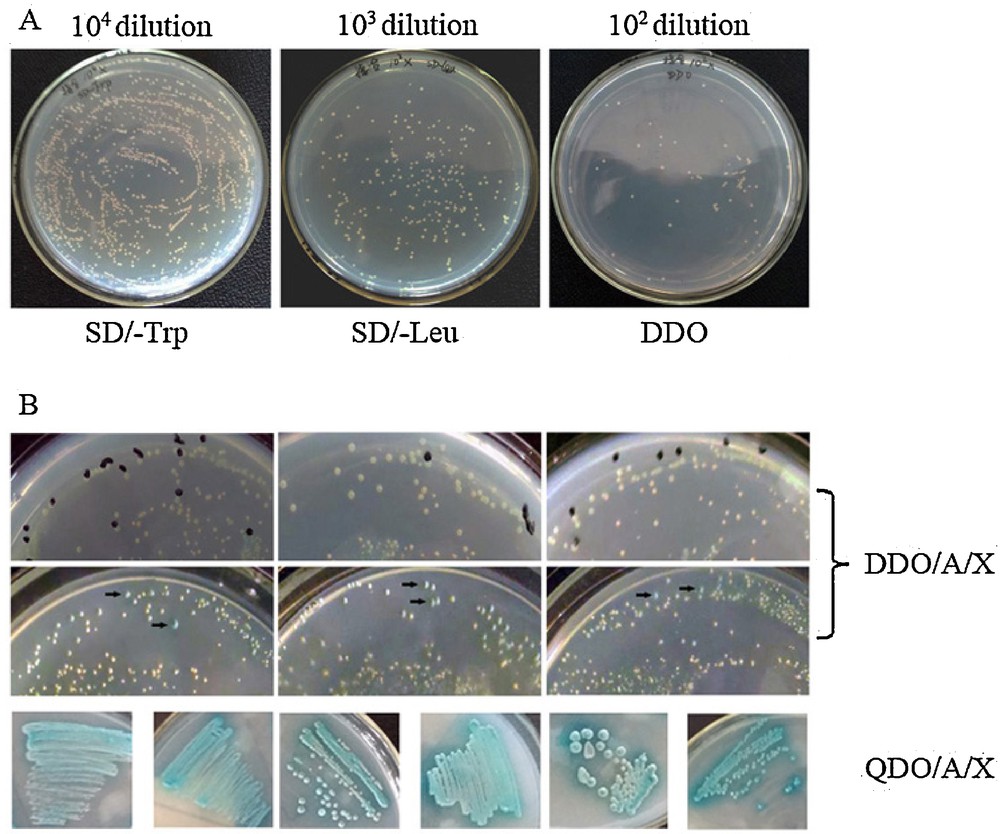
Library screening. (A) Mating efficiency. ∼1500 clonies grew on the 104 dilution SD/-Trp plate. ∼260 clonies grew on the 103 dilution SD/-Leu plate. ∼85 colonies grew on the 102 dilution DDO plate. Mating efficiency was 3.27% (2% ≥ standard ≥ 5%). (B) Representative colonies showing positive interactions visualized as blue on DDO/A/X plates during the first screening (black arrow). Blue colonies from DDO/A/X were plated onto QDO/A/X for more stringent screening.
3.2 The analysis of bovineCMYA1/Xin repeats conservation
Previously, we obtained the bovine CMYA1 protein partial sequence that is 1,132 aa with a predicted molecular mass of 124 kDa (GenBank: ADM45307.1). Conservation analysis was performed using BLAST on NCBI, and we found that the bovine CMYA1 protein is 73% identical (81% identity within the Xin repeat region) to the corresponding human protein (1,121 amino acids, 122 kDa, Sequence ID: NP_919269.2). Bovine CMYA1 also shares 80% identity within the Xin repeat region of mouse (Sequence ID: NP_035854.3). Whereas nearly 50% of the CMYA1N-terminus consists of 14 Xin repeats (Fig. 3A), there is only one Xin-repeat in C-terminus of CMYA1. Because it has been shown that the minimal number of Xin-repeats that have function is at least 3 [17], there is little chance that the one Xin-repeat in C-terminus is functional. Therefore, this one Xin-repeat was not targeted in this research. The consensus sequence of Xin-repeats was GDV(K/Q/R/S)XX(R/K/T)WLFET(Q/R/K/T)PLD (Fig. 3B).
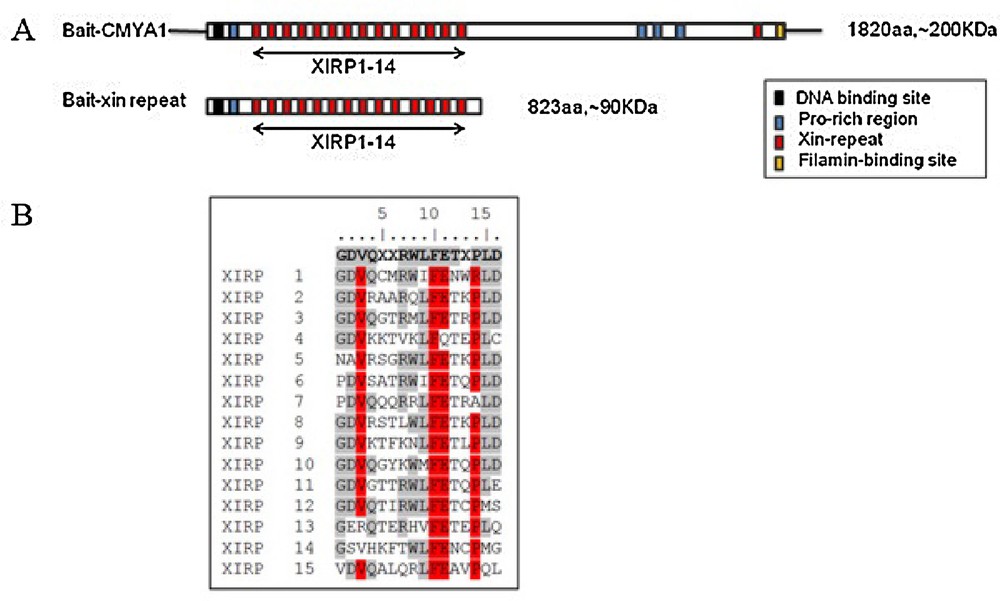
Characterization of bovine CMYA1. (A) Schematic representation of the domain structures of bovine CMYA1. Xin repeats are indicated by red boxes. Amino acid (aa) residues for the respective constructs are indicated. CMYA1 variants and the baits that were constructed for use in the yeast two-hybrid screen. (B) The sequence of the 15 bovine Xin repeats and their 16 aa consensus sequence (top line, boldface). Identical amino acids at each corresponding position are shown in grey and highly conserved residues are shown in red. Masquer
Characterization of bovine CMYA1. (A) Schematic representation of the domain structures of bovine CMYA1. Xin repeats are indicated by red boxes. Amino acid (aa) residues for the respective constructs are indicated. CMYA1 variants and the baits that were constructed for ... Lire la suite
3.3 Potential CMYA1/Xin repeats interacting proteins
To identify potential Xin-repeat binding proteins in bovine skeletal muscle, a yeast two-hybrid screen was performed on a bovine muscle cDNA library using full-length CMYA1 and Xin-repeats as the bait (Fig. 4A). CMYA1/Xin-repeats do not cause autoactivation or toxicity when introduced in yeast (Fig. 4C). A total of 2.8 million independent clones were screened producing 670 clones that interact with the full-length CMYA1 bait and 900 clones that interact with the Xin-repeats bait (Fig. 5B, Table 1). Positive clones were PCR-amplified, and inserts that were larger than 300 bp were selected for sequencing. After performing the screen twice, a total of 27 putative interacting proteins were identified (Table 3). There were 14 proteins that interacted with CMYA1, 16 proteins that interacted with the Xin-repeats, and three proteins (RPL35A, RPL21 and EIF3G) that interacted with both CMYA1 and Xin-repeats making them candidate interacting proteins (Table 3).
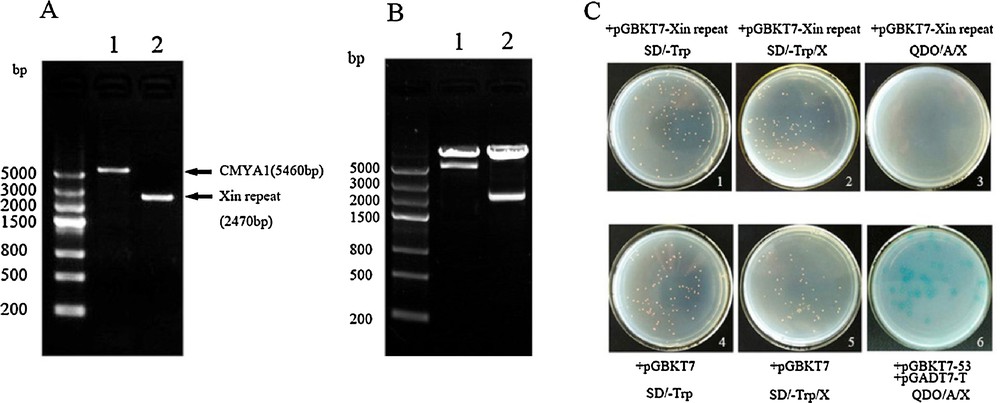
(A) The fragment of CMYA1and the Xin repeats. 1: CMYA1fragment (1–1820aa) that contains the complete CDS. 2: The Xin repeats domain of CMYA1(1–823aa). (B) Restriction enzyme validation of the constructed bait vectors pGBKT7-(CMYA1/Xin repeat). 1: CMYA1 bait vector digested with EcoRI and SalI. 2: Xin repeat bait vector digested with EcoRI and BamHI. (C) Transformation of pGBKT7-Xin repeats into the Y2H Gold strain, autoactivation and toxicity test of the bait vectors. (1, 4) The strain transformed with pGBKT7-Xin repeats shows no toxicity. (2, 3) The strain transformed with pGBKT7-Xin repeats shows no autoactivation. (5, 6) The strain transformed with pGBKT7 and plated on SD/-Trp/X and the strain co-transformed with pGBKT7–53/pGADT7-T as a control (The result of bait pGBKT7-CMYA1 was similar to theXin repeats, data not shown). Masquer
(A) The fragment of CMYA1and the Xin repeats. 1: CMYA1fragment (1–1820aa) that contains the complete CDS. 2: The Xin repeats domain of CMYA1(1–823aa). (B) Restriction enzyme validation of the constructed bait vectors pGBKT7-(CMYA1/Xin repeat). 1: CMYA1 bait vector digested with ... Lire la suite
The BLAST results of positive interacting proteins from the CMYA1/Xin repeats bait.
| No. | Genbank accession No. | Bovine prey genes (GenBank accession number) | Baits (CMYA1/Xin repeat) |
| 1 | NM_001013590.2 | Tropomyosin 1 (alpha) (TPM1) | CMYA1 |
| 2 | NM_001077877.2 | Calsequestrin 1 (fast-twitch, skeletal muscle) (CASQ1) | Xin repeat |
| 3 | NM_001079578.1 | Myosin, light chain 1, alkali; skeletal, fast (MYL1) | Xin repeat |
| 4 | NM_001046247.2 | Laminin, beta 2 (laminin S) (LAMB2) | Xin repeat |
| 5 | NM_001075647.1 | Myosin light chain, phosphorylatable, fast skeletal muscle (MYLPF) | Xin repeat |
| 6 | NM_001014915.1 | Titin-cap (telethonin) (TCAP) | CMYA1 |
| 7 | NM_001083480.2 | Actin filament (CAPZA1) | Xin repeat |
| 8 | NM_001035445.2 | Ribosomal protein S4, X-linked (RPS4 X) | CMYA1 |
| 9 | XM_005902269.1 | Ribosomal protein S6 (RPS6) | CMYA1 |
| 10 | BC148016.1 | Ribosomal protein S9 | Xin repeat |
| 11 | XM_005213762.2 | Ribosomal protein L21 (RPL21), transcript variant X2 | CMYA1 |
| 12 | NR_036642.1 | 18S ribosomal RNA (RN18S1), ribosomal RNA | CMYA1 |
| 13 | NM_001025321.3 | Ribosomal protein L35a ( RPL35A ) | CMYA1/Xin repeat |
| 14 | NM_001191412.1 | Bos taurus ribosomal protein L21 ( RPL21 ) | CMYA1/Xin repeat |
| 15 | NM_001079623.2 | Eukaryotic translation initiation factor 3, subunit G ( EIF3G ) | CMYA1/Xin repeat |
| 16 | XM_005204863.1 | Splicing factor proline/glutamine-rich (SFPQ), transcript variant X2 | CMYA1 |
| 17 | XM_005893725.1 | Delta-like 1 homolog (Drosophila) (DLK1) | CMYA1 |
| 18 | NM_001101075.2 | Alanyl-tRNA synthetase (AARS) | Xin repeat |
| 19 | XM_005210737.1 | Family with sequence similarity 184, member A (FAM184A), transcript variant X5 | CMYA1 |
| 20 | NM_001075424.1 | Mevalonate (diphospho) decarboxylase (MVD) | CMYA1 |
| 21 | NM_001102041.2 | KIAA0226-like ortholog (KIAA0226L) | Xin repeat |
| 22 | XM_005227674.1 | Transketolase-like 1 (TKTL1), transcript variant X2 | CMYA1 |
| 23 | NM_001103311.1 | Melanoma antigen family D, 4B (MAGED4B) | Xin repeat |
| 24 | NM_001076903.1 | Phosphoglucomutase 1 (PGM1) | Xin repeat |
| 25 | BN001315.1 | MUC16 gene for mucin-16, exons 1–52 | Xin repeat |
| 26 | NM_001034034.2 | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Xin repeat |
| 27 | NM_001076339.1 | THAP domain containing 5 (THAP5) | Xin repeat |
3.4 Verification of candidate CMYA1/Xin-repeats interacting proteins
To determine if the three candidates (RPL35A, RPL21 and EIF3G) were CMYA1/Xin-repeats interacting proteins, the full-length CDS of each candidate was cloned into the pGADT7 prey vector to be used for a mating experiment (Fig. 6). One-to-one Y2H assays were performed between the three prey candidates and the two baits (CMYA1/Xin repeats). Notably, RPL35A, RPL21 and EIF3G all interacted with both CMYA1 and the Xin-repeats, which were also in accordance with the results from when the library was screened previously (Fig. 7).
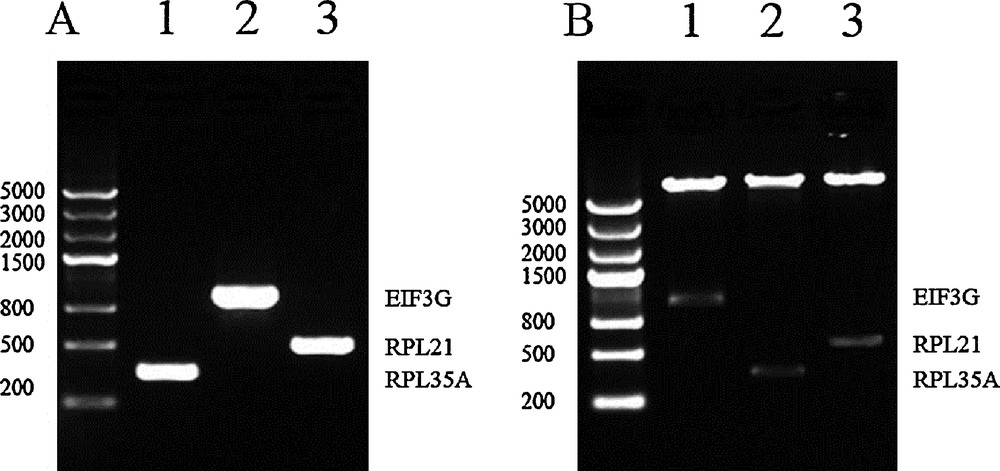
Agarose gel analysis of the three interacting proteins identified from both screens. (A) PCR-amplification of the complete CDs of the three prey genes. 1: RPL35 A was 333 bp. 2: EIF3G was 963 bp. 3: RPL21 was 483 bp. (B) Dual-enzyme digestion identified the constructed vectors of pGADT7-prey. 1: prey EIF3 G was digested by NdeI and EcoRI. 2: prey RPL35 A was digested by NdeI and BamHI. 3: RPL21 was digested by NdeI and EcoRI.
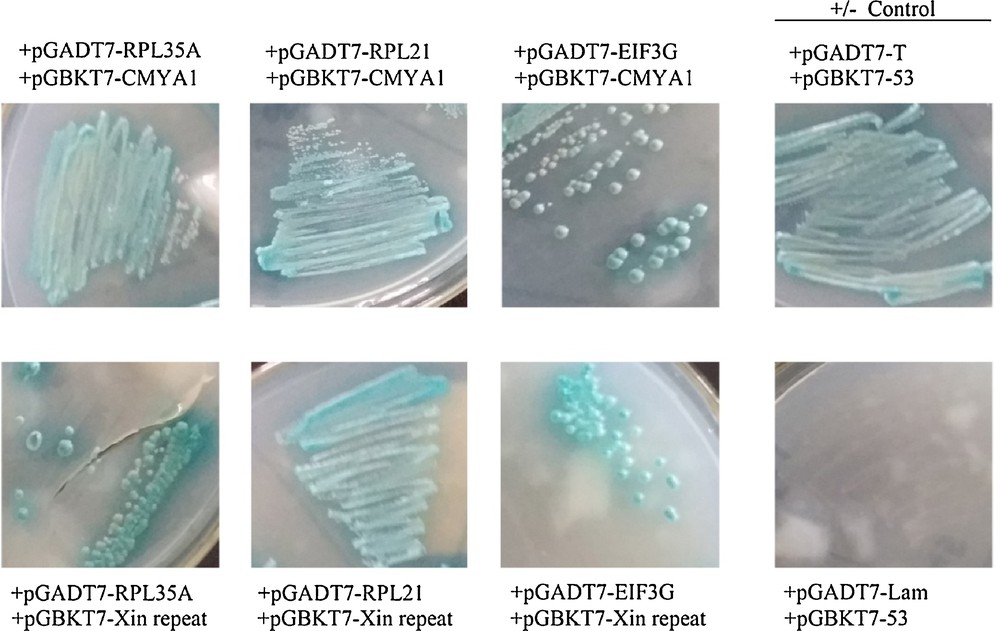
One-to-one assay of candidate interacting proteins. Co-culture of the yeast Y187 strain containing candidate AD-prey vectors and the Y2H strain containing BD-CMYA1 or BD-Xin repeat vectors, respectively, on QDO/X/A 3–5 days at 30 °C. The yeast strains contained pGADT7-T and pGBKT7–53 as a positive control, and pGADT7-T and pGBKT7-Lam as a negative control. Blue clonies that grow on QDO/X/A show that the bait protein can bind prey proteins; conversely, no clonies indicate no-interactions between bait and prey.
4 Discussion
The yeast two-hybrid system is a highly feasible approach to identify interacting proteins. In this study, a high-quality cDNA library of bovine muscle was generated for Y2H and applied to study proteins that function during bovine muscle development. The cDNA library was screened twice using the full-length CMYA1 protein and a part of it that contained the Xin-repeats, and 27 putative interacting proteins were identified. Furthermore, most of the interacting proteins have not been reported before. The actin-based protein that was previously reported to interact with CMYA1was also identified in our study, indicating that our library is dependable. In the future, the cDNA library can also be used to detect the interacting proteins of other CMYA1 family members and other Xin-repeat containing proteins.
The 27 candidate-interacting proteins were generally classified into actin cytoskeleton, replication/transcription/translation, and cell signaling three categories according to their biological functions. The actin cytoskeleton is a complex protein network including MYL1, TPM1, MYLPF, and CAPZA1. The biological process of actin cytoskeleton is responsible for muscle contraction, translation, muscle organ development and striated muscle tissue development etc. excepted for keeping cell structure and shape and providing cell motility [18–21]. Replication/transcription/translation is the intracellular activity in which the DNA is transcribed to RNA and the RNA is translated, via the ribosome, into protein [22]. Cell signaling is involved in many complex cell actions and cellular activities [19]. All of these cellular activities are associated with muscle development. The identification of 27 putative interacting proteins of CMYA1 and the Xin repeats indicates that CMYA1/Xin-repeat may play a role in muscle development mechanism by participating with other pathways. The putative interacting proteins from this study provided many new targets for further investigation of the functions of CMYA1/Xin-repeats. From the Y2H screening results, three proteins (RPL35A, RPL21 and EIF3G) interacted with both CMYA1 and the Xin-repeats. This finding provides further proof that the Xin-repeats is a vital domain of CMYA1 that is required for interaction with other proteins and that it will be important to explore the potential function of the Xin-repeats domain.
RPL35A and RPL21 identified in this study are ribosome proteins that are highly abundant and universally found in all animals. Most ribosome proteins participate in ribosome biogenesis; however, a number have independent functions, such as interactions with other proteins [22]. It was originally thought that translation of select transcripts occurred at the cytoskeleton [18]; however, new evidence indicates that a large portion of translation may occur at ribosomes that are bound to the cytoskeleton [22]. Electron microscopy has shown that ribosomes co-localize with components of the cytoskeleton [23]. Several published reports have implicated actin [24], intermediate filaments [25] and tubulin [26,27] as the cytoskeletal components responsible for ribosomal interactions. Altering the balance between the protein synthesis and degradation could regulate the skeletal muscle development [28], but the mechanism of how protein synthesis can affect muscle mass is unclear [29]. So, the interaction between RPL35A and CMYA1/Xin repeats and RPL21 and CMYA1/Xin-repeats provides a new target for research on the mechanism of how ribosome biogenesis may influence muscle mass. Finally, CMYA1 was reported to co-localize with the cytoskeleton, the Xin-repeats can also bind actin, and Xin-repeats interact with RPL35A and RPL21; therefore, proteins that contain Xin-repeats may play a role in translation, and this role may be mediated through interactions of the ribosome with components of the cytoskeleton.
EIF3G is a protein component of the eukaryotic initiation factor that was detected in this study. The eukaryotic initiation factor is a key regulator of muscle development and protein synthesis [30]. The EIF3 complex is required for several steps in the initiation of protein synthesis [31–33], while EIF3 was also reported to have a specific effect on myofibrillar protein synthesis [34]. It has been suggested that muscle EIF3 can associate with specific proteins and play a role in mRNA activation and translation during muscle differentiation [30,35,36]. EIF3G, identified in this study, is a subunit of EIF3. EIF4 is another member of the eukaryotic initiation factors. EIF4EBP binds toEIF4E, and together, they can repress translation. It has been reported that EIF4EBP can regulate the translation and level of Mef2 protein [37]. CMYA1 is a downstream target of transcription factors Nkx2.5 and Mef2 in the BMP-Nkx2.5-Mef2C pathway [1,38]. These results suggest that CMYA1/Xin-repeats is EIF3G binding protein, which may provide a mechanism of how CMYA1 and the Xin-repeats regulate the BMP-Nkx2.5-Mef2C pathway.
5 Conclusions
To investigate the proteins that interact with the Xin-repeats region of CMYA1, we generated a high-quality bovine muscle cDNA library and two bait constructs (CMYA1/Xin repeat) for use with the Y2H system. Following two screenings of the cDNA library, we found 27 proteins that interacted with CMYA1/Xin-repeats, three of which (RPL35A, RPL21 and EIF3G) interacted with both the CMYA1 bait and the Xin-repeats bait. CMYA1 interacting proteins that were previously reported, such as the actin-based protein that was also found in our study, indicate that our Y2H system is dependable. The three proteins that interacted with CMYA1/Xin-repeats provide new insights on how protein synthesis and eukaryotic initiation can influence muscle development. In conclusion, this study provides new targets for further investigation of the functions of CMYA1 and other proteins that contain Xin-repeats.
Ethics approval statement
All animal sample procedures were approved and performed in accordance with institutional guidelines of Tianjin Agriculture University.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Data citation
Not applicable.
Authors’ contributions
Xiangbo Xin and Ting Wang contributed equally to this study. Xinfeng Liu reviewed the manuscript, Guoning Sui and Congfei Jin collected samples of bovine, Yingwei Yue, Shuping Yang and Xiangbo Xin GZ contributed in lab analysis and maintained the database of the project. All authors read and approved the final manuscript.
Funding information
1. TianJin Program on Key Basic Research Projects (11JCZDJC17600); 2. National Science and Technology Major Project (2014ZX08007–002).
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
This work was accomplished in the Tianjin International Joint Research and Development Center.
The authors thank the Inner Mongolia University bovine breeding bases provided samples of Simmental cattle. Thanks are also due to GuangPeng Li professor and his laboratory provided technical guidance of collecting samples.


