1 Introduction
For most, mangroves simply represent large areas of trees that mainly occur in the intertidal zone, between land and sea, in the tropics. On the marine border, this ecosystem is characterized by “normal” seawater with pH, temperature, salinity, and dissolved oxygen similar to that of other tropical marine environments. Most studies have focused on the terrestrial zone of the mangrove, and very few have analyzed the prokaryotes colonizing marine sediments in this ecosystem. Microbial mats in mangrove tidal channels often have an outer layer of cyanobacteria and inner layers composed of anoxygenic phototrophic bacteria [1,2].
Bacterial cells colonizing aquatic sediments are generally organized, based on their physiology, in a vertical axis forming a laminated structure. The top layer is usually dominated by a filamentous cyanobacteria species and benthic diatoms followed by a distinct layer of anoxygenic phototrophs (purple and green bacteria). White patches of filamentous colorless sulfur-oxidizing bacteria, belonging to Beggiatoaceae, can occasionally be observed at the surface of the marine sediment [3,4]. These first upper layers are followed by sulfate-reducing bacteria, which are responsible for producing sulfides under anaerobic conditions, which can reach up to 20 mM in marine mangrove sediments [5].
Anoxygenic photolithotrophic bacteria belong to the categories of green sulfur bacteria (GSB) and purple sulfur bacteria (PSB), which can form clearly visible colored microbial mats in various natural and/or anthropogenically influenced environments [6]. Purple bacteria can also be permanently exposed to oxygen at the sediment surface, as observed during mass development of PSB in intertidal zones [7–10].
PSB in anaerobic aquatic environments perform anaerobic and anoxygenic photosynthesis, in contrast to cyanobacteria [11]. They do not use water as a reducing agent and thus do not produce oxygen. They oxidize hydrogen sulfide (H2S) or sulfides (HS−) to produce internal or external granules of elemental sulfur (S0), depending on the bacterial species concerned [12]. Moreover, these bacteria contain a single photosystem that is structurally related to PSII in cyanobacteria and chloroplasts. Phototrophic purple bacteria are the most phylogenetically diverse anoxyphototrophs, with members in the α, β, and γ classes of the Proteobacteria [13,14]. Functional genes, such as the M subunit of the photosynthetic reaction center, can be used to confirm classical phylogenetic trees obtained from 16S rRNA sequences. PSB strains may be non-cultivable and molecular analyses can be complemented by ultrastructural analyses coupled with X-ray analysis to detect the elemental sulfur inside the cells [15].
PSB belong to the order Chromatiales (class Gammaproteobacteria), which contains four families, two of them are phototrophic: the Chromatiaceae and the Ectothiorhodospiraceae. The major phenotypic difference between these two families concerns the localization of sulfur, internal versus external sulfur granules, respectively. The members of Chromatiaceae belong to three phylogenetic groups: (1) marine and halophilic species comprising the genera Halochromatium, Isochromatium, and Marichromatium (2) freshwater species belonging to the genera Chromatium and Thiocystis, and (3) water/mud from freshwater to marine species of the genera Thiocapsa and Amoebobacter [16].
Anoxygenic photolithotrophic bacteria use reduced sulfur species as an electron source [17] to grow. Thus, the sulfides, produced in the mangrove sediments by sulfate-reducing bacteria (SRB), diffuse upward into the bottom waters and can be used by anoxygenic phototrophic bacteria. In mangroves, anoxygenic photosynthetic bacteria, including purple sulfur bacteria (Chromatium spp.) and purple non-sulfur bacteria (Rhodopseudomonas spp.), have been isolated from terrestrial mangrove soils in Pichavaram, South India [18–20]. However, there is no description available from the literature concerning PSB in marine mangrove sediments.
Here, we explored the contribution of PSB to marine mangrove microbial communities using phylogenetic analyses (the 16S rRNA gene and the PufM subunit of the photosynthetic reaction center) coupled with ultrastructural investigations (ESEM, TEM, EDXS) and FISH experiments to characterize and identify these dominant anoxygenic phototrophic bacteria from this tropical marine environment.
2 Materials and methods
2.1 Sampling site
Samples were manually collected from a depth of approximately 1 m, using a syringe, from the periphyton of a tropical marine mangrove in Guadeloupe (Caribbean) under a Rhizophora mangle canopy. Samples were collected at two sites (N16°16′560″/W61°33′303″ and N16°16′826″/W61°33′220″) recently investigated to characterize the sediment chemistry [21,22]. Temperature and salinity at a depth of 1 m were relatively constant with average values of 28 °C and 35‰, respectively. After collection, samples were immediately transported to the laboratory and observed and photographed with a stereomicroscope. These bacterial cells can be easily concentrated under a spot light as they migrate as far as possible from the strong light source due to their negative phototactic behavior (Fig. 1c). Such “purified samples” were used for all experiments presented in this study.
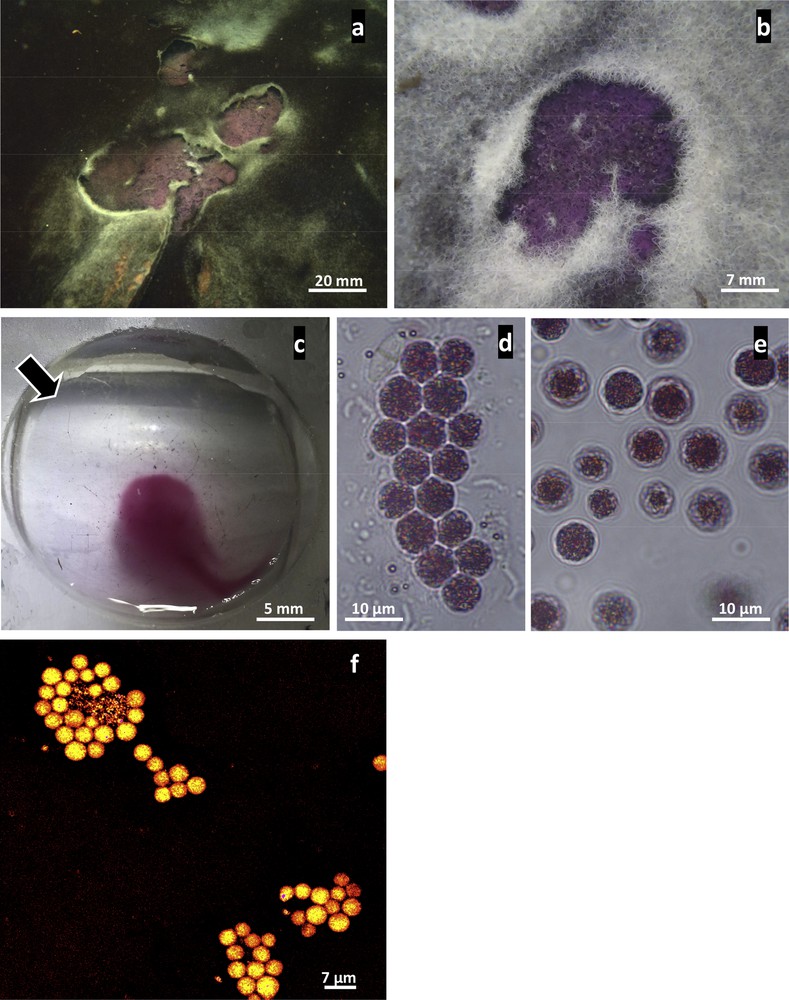
Description of purple mat of PSB novel phenotype from marine mangrove (a–f: light micrographs). a, b: In situ images showing purple mats observed either within black mats mostly composed of cyanobacterial species belonging to the genus Planktotrichoides (a) or filamentous white mats of sulfur-oxidizing Beggiatoa (b). c: This pigmented mat appears to be composed almost exclusively of a dense population of purple cells with clear negative phototactic behavior. In the presence of a light source (arrow), the cells moved to the opposite side, allowing their easy enrichment from sediment particles and micro-eukaryotic organisms inhabiting the sediment. d, e: Light micrographs showing single ovoid cells that appear purple due to the presence of pigment within the cell. Moreover, small refractive internal granules are clearly visible in each cell. These unicellular organisms generally form aggregate structures typical of this bacterial species (e). f: FISH experiments showing strong hybridization with the specific probe designed in this study (probe Hc48f Cy3 dye labelled).
2.2 Analysis of the photosynthetic pigments.
Absorption spectra (λ between 400 nm and 900 nm) were obtained from intact cells suspended in 30% bovine serum albumin [23] or after pigment extraction for 30 min in the dark at −20 °C in an acetone/methanol solution (7:2, vol/vol). Spectra were obtained using a SECONAM spectrophotometer.
2.3 Scanning electron microscopy
Samples for SEM observations were fixed for 2 h at 4 °C in a 2.5% glutaraldehyde solution in cacodylate buffer (900 mOsM, pH 7.2). Then, they were dehydrated in graded concentrations of acetone, critical point dried in CO2, and sputter-coated with gold before observation with a FEI Quanta 250 at 20 kV.
2.4 Energy-dispersive X-ray spectroscopy (EDXs) analysis
Freshly fixed samples (as described for SEM preparation samples above) were observed using an environmental scanning electronic microscope (FEI Quanta 250) operating at 15 kV under an environmental pressure of 7 Torr at 5 °C to detect elemental compounds of individuals. Just before observation, samples were quickly rinsed in deionized water to remove salts. EDX spectra were obtained using an M-max 50-mm2 Oxford detector.
2.5 Transmission electron microscopy (TEM)
Freshly collected samples were prepared for TEM as previously described [24]. Two grids containing four to five ultrathin sections (60 nm in thickness) were observed using a FEI TecnaiG2 at 200 kV.
2.6 PCR amplification and DNA sequencing
Bacteria were concentrated after exposure to light (Fig. 1c) and centrifuged at 6000 g for 3 min. The resulting pellet was suspended in 0.22-μm-filtered sterile seawater and washed three times. DNA was extracted from the last pellet obtained using the DNEasy extraction kit (Qiagen) according to the manufacturer's protocol. Genes encoding bacterial 16S rRNA were amplified using the universal primer set (8F-1492R) as described previously [25]. Molecular analysis, based on the pufM gene, encoding the M-subunit of the photosynthetic reaction center, was performed using the primer set (PufMF–PufWAW) [14,26]. Sanger sequencing of the PCR products (16S rDNA and pufM genes) was realized by GATC (http://www.gatc-biotech.com).
2.7 Phylogenetic analysis
The obtained 16S rRNA gene sequence was compared with the National Center of Biotechnology information (NCBI) (http://www.ncbi.nlm.nih.gov) database. Nearest relatives and sequence similarities were determined by BLAST search [27]). 16S rDNA sequences of representative species of the Chromatiaceae were aligned using the Clustal X program [28] and refined manually. Phylogenetic analyses were performed using MEGA version 5.2 software [29]. The phylogenetic trees were inferred by the maximum-likelihood method (ML) [30]. Node robustness was assessed by performing 1000 bootstrap replicates [31] and only bootstrap values above 50% are indicated at the nodes of the trees. Chloroflexus aurantiacus strain DSM 637 gi|15551748|emb|AJ308501.1|) was used as an outgroup. Following analysis, the 16S rDNA partial sequence obtained in this study was deposited in the GenBank database under the accession number KX714823.
2.8 Fluorescent in situ hybridization (FISH) experiments
Bacteria were fixed for 2–3 h at 4 °C in 4% paraformaldehyde in seawater. Specimens were then washed three times for 10 min each at 4 °C in seawater and stored in 70% ethanol at 4 °C until analysis. Two oligonucleotide probes (Cy3 labelled) were used: Eub338 (5’-GCTGCCTCCCGTAGGAGT-3’), targeting most members of the eubacteria [32], and a more specific probe (Hc48f), designed in this study according to the 16S rDNA sequence obtained. The probe Hc48f (5’-CCGTCCGCCACTCTACTT-3’) was checked against 16S rRNA-encoding Chromatiales sequences in RDP using the implemented tool Probe Match [33]. The Hc48f probe hybridized in silico with uncultured bacteria belonging to the genera Halochromatium (4/537 sequences), Thioflavicoccus (3/74 sequences), and Thiohalocapsa (0/795 sequences), with no mismatches.
Hybridization experiments were similar to those previously described [34], except that Hc48f was used with a hybridization buffer containing 40% formamide. The environmental PSB novel phenotype investigated here showed no auto-fluorescence with the Cy3 filter used.
3 Results
We detected small purple areas in marine mangrove environments close to Rhizophora roots, either within cyanobacterial mats (Fig. 1a), containing mostly Planktotrichoides species [35], or white bacterial mats (Fig. 1b) composed of large filaments of Beggiatoaceae [4]. In both cases, these purple mats were localized at the interface between anaerobic sediments and oxygenated seawater environments. The purple mats were composed almost exclusively of a dense population (Figs. 1c–d) of highly motile, purple cells (Fig. 1c). These bacterial cells are obviously negatively phototactic in the lab under high light intensity, but according to the ecological niche of PSB in the environment, a positive phototactic response would be expected in low light intensity in order to find the good light intensity for their photosynthesis. Bright field microscopy revealed the presence of refractive granules (Figs. 1d–e) randomly distributed in spherically-shaped cells with a mean diameter of 6.75 ± 0.95 μm. Moreover, each individual cell was purple-red, reflecting its pigment composition (Figs. 1d–e), responsible for the microbial-mat color observed in the field (Figs. 1a–b). These unicellular micro-organisms could form irregular clumps of cells (Fig. 1d).
Spectra obtained from the “enriched fractions” showed that the major photosynthetic pigment present was bacteriochlorophyll a. This photosynthetic pigment is characterized by a main peak at 850 nm in living cells suspended in 30% bovine serum albumin and 770 nm for acetone-extracted pigments (Fig. 2). Bacteriochlorophyll a is the most widespread pigment in PSB even if not all species show a maximum for the bacteriochlorophyll a in living cells at 850 nm like for spirriloxanthin-containing species (two maxima at 805 and 870 nm) or okenone-containing species (a single peak at 830 nm) [36]. Thus, these data strongly suggest that the strain analyzed here belongs to PSB.
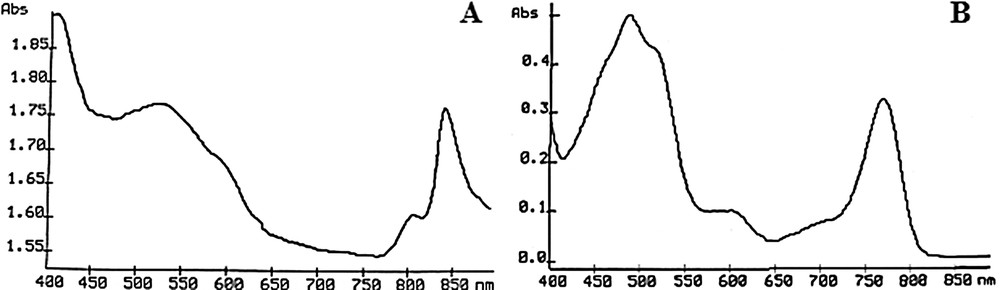
Pigment analysis. Absorption spectra obtained from a purified fraction of purple bacterial cells in 30% BSA solution (A) and after extraction of the pigment with an acetone/methanol solution (B). The main peaks obtained at 850 nm (A) and 770 nm (B) demonstrate the presence of bacteriochlorophyll a as the major photosynthetic pigment within these purple cells.
SEM observations showed that these micro-organisms occur singly and are ovoid with a monopolar tuft of flagella (Figs. 3a–c). TEM showed a double membrane typical of Gram negative bacterial cell wall (Figs. 3d–e). Moreover, cells contained intracytoplasmic vesicular membrane systems extending throughout the cell in which the photosynthetic pigments are located. No gas vesicles could be detected. The “large empty vesicles” observed inside the cytoplasmic volume presumably represent the location of sulfur inclusions lost during the dehydration and embedding processes. Freshly collected samples were analyzed using an ESEM microscope coupled with an EDX detector to identify chemical content. The white dots observed in each cell indicated internal compounds with higher atomic density (Fig. 4a) than the cellular compounds. EDX spectra obtained from several individuals clearly showed a major peak for sulfur (Fig. 4b), suggesting elemental sulfur granules. Small peaks of iron and chrome were detected (Figs. 4b–c), independent of bacterial cells (Fig. 4g), reflecting the underlying specimen holder, as suggested by the elemental map (Figs. 4c–i). Sulfur was strongly detected and concerned the whole cell (Fig. 4h), as shown by the sulfur distribution and mix-map picture obtained for this element (Fig. 4i). These results suggest that elemental sulfur is formed as an intermediate oxidation product and stored as refractive globules easily detectable by light microscopy (Fig. 1d). Such phenotypic properties would thus assign this bacterial species to phototrophic PSB. Indeed, bacteria are involved in the sulfur cycle and these could be phototrophic sulfur bacteria, given their purple color and the presence of bacteriochlorophyll a. However, the few attempts to culture such bacteria in liquid media (under aerobic or anaerobic conditions) have failed [37], making their characterization difficult. Indeed, Halochromatium-like PSB from Guadeloupe was unable to grow photo-lithoautotrophically [anaerobic, aerobic, daylight exposure, Na2S.9H2O (1 mM), Na2S2O3·5H2O (5 mM) and NaHCO3 (0.1% w/v)] or photo-organotrophically (using citrate or glucose).
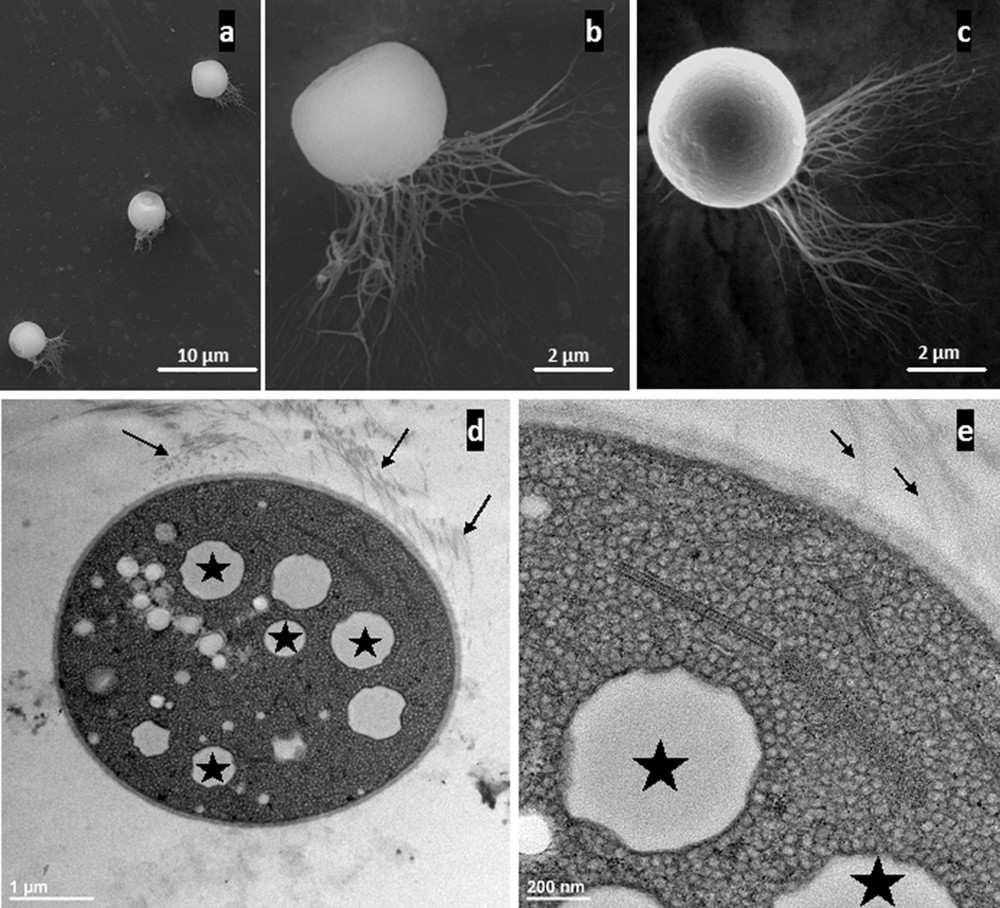
Ultrastructural analysis of freshly collected individuals from marine mangrove (a–c: SEM micrographs; d–e: TEM micrographs). a–c: Low magnification of three cells showing the ovoid shape of this bacterial phenotype. Two important features are clear: the presence of numerous flagella (b, c), showing that these bacteria are motile by means of lophotrichous flagella, and flattened at the opposite pole (a, c). d: Transverse section of one bacterial cell. The cytoplasmic volume appears to be filled with small vesicles corresponding to internal photosynthetic membrane systems typical of PSB ultrastructure. Moreover, larger white “empty vesicles”, which are probably former sites of sulfur granules (stars), represent the second main component of the cytoplasmic volume. Lophotrichous flagella (small arrows) are located on one side of the bacteria. e: Higher magnification, focusing on the density of the intracytoplasmic vesicular membrane systems (∼20 nm in thickness) typical of bacterial species belonging to the Chromatiaceae. The photosynthetic vesicular membrane systems extend throughout the cell, occupying most of the cytoplasm around the sulfur granules (stars). Flagella are indicated by the small arrows.
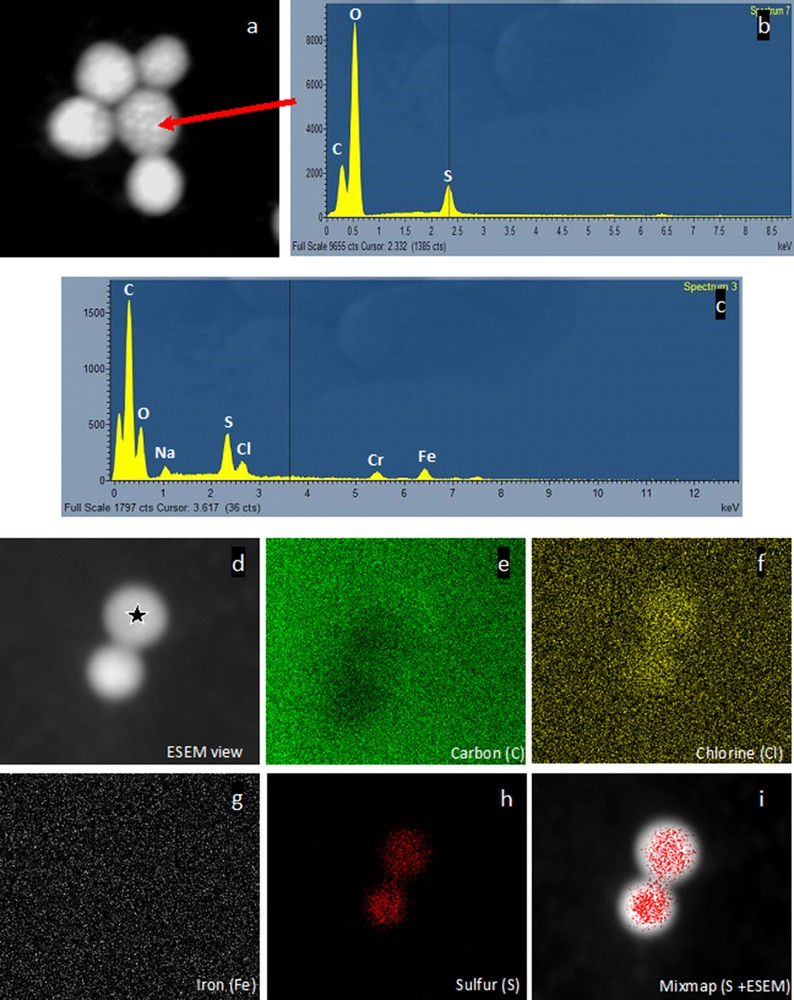
Environmental Scanning Electron Microscopy images and analyses recorded on lightly fixed individuals under a 650-mPa water vapor atmosphere at 15 kV (a: backscattered electron image: b, c: EDXS spectra, d–i: X ray maps characterizing distributions of various chemical elements within the sample). a: Image obtained using a secondary electron detector with a 15-kV accelerating voltage. The image is dominated by backscattered electrons due to the high penetration power of the incident electrons in the specimen. The resulting atomic number (Z) contrast reveals high-Z-number granules (sulfur) as bright areas. Secondary contrast is present, but only visible at the periphery of the cell. b: The EDX spectrum obtained from one single bright granule (arrow) clearly indicates that the main element present in the cytoplasmic granule is sulfur. Thus, elemental sulfur accumulates as globules inside the cells from sulfides under anaerobic culture conditions. c: EDX spectrum obtained from one of the cells (star) seen in 4b. Elemental sodium, sulfur, and chlorine are detected in the bacterial cells, whereas chromium and iron probably come from the specimen holder, as demonstrated by mapping (d–i). d–i: Chlorine and sulfur are only located within the bacterial cell (f, h), whereas carbon (e) is present everywhere (biological sample and specimen holder). However, iron clearly comes from the specimen holder (g), as no iron can be located within the bacterial cell analyzed (g). The sulfur map (h) clearly demonstrates that sulfur is located inside the bacteria as confirmed by the mix-map image (i).
In order to ascertain the phylogenetic positions of this purple bacterial isolate, we amplified and partially sequenced the 16S rRNA gene (848 bp fragment, accession number KX714823). Phylogenetic analyses revealed that this environmental PSB novel phenotype belongs to the family Chromatiaceae and is related to a group of marine and halophilic species genera including Halochromatium and Thiohalocapsa (Fig. 5). The highest 16S rRNA gene sequence similarity was shared with Halochromatium, Hch. salexigens 6310 (93%) and Hch. glycolicum BN 3201 (93%). According to Yarza et al. [38], such similarity suggests that these strains, defined as “Halochromatium”, may actually belong to at least two different genera. Additional studies will be required to accurately determine the taxonomy of our isolate. The phylogenetic data were verified by fluorescence in situ hybridization (FISH) with a specific probe designed for FISH (Fig. 1f). The strong hybridization signals obtained with either EUB 338 (not shown) or Hc48f (Fig. 1f), strongly suggest that the 16S rDNA sequence obtained came from these spherical cells and not contaminants colonizing the sediment. We further pursued the phylogenetic analysis by amplifying and analyzing the metabolic gene pufM [13,39]. The pufM gene, encoding the medium (M) subunit of the photosynthetic reaction center of anoxygenic photosynthetic bacteria, is universally distributed across purple phototrophic bacteria [40,41]. The PCR amplified band was of the expected size (∼245 bp) for the pufM primer set used, but gave only a 165 bp sequence available for phylogenetic analysis. This small pufM sequence shared 78% identity with Halochromatium sp. and Thiorhodovibrio sp., and 79% identity with Thiohalocapsa sp. We then converted the nucleotide sequences of the pufM genes into amino acid sequences and compared them with available sequences to confirm these results. The PufM sequence was still distinct from all available sequences and most closely related to Halochromatium sp. (93% identity) and Thiorhodovibrio sp. (94% identity). Moreover, the maximum-likelihood phylogenetic tree based on M subunit sequences (Fig. 6) suggests (even if some node values are weak) that this mangrove isolate diverged independently from the evolution of the Halochromatium and Thiorhodovibrio lineages.
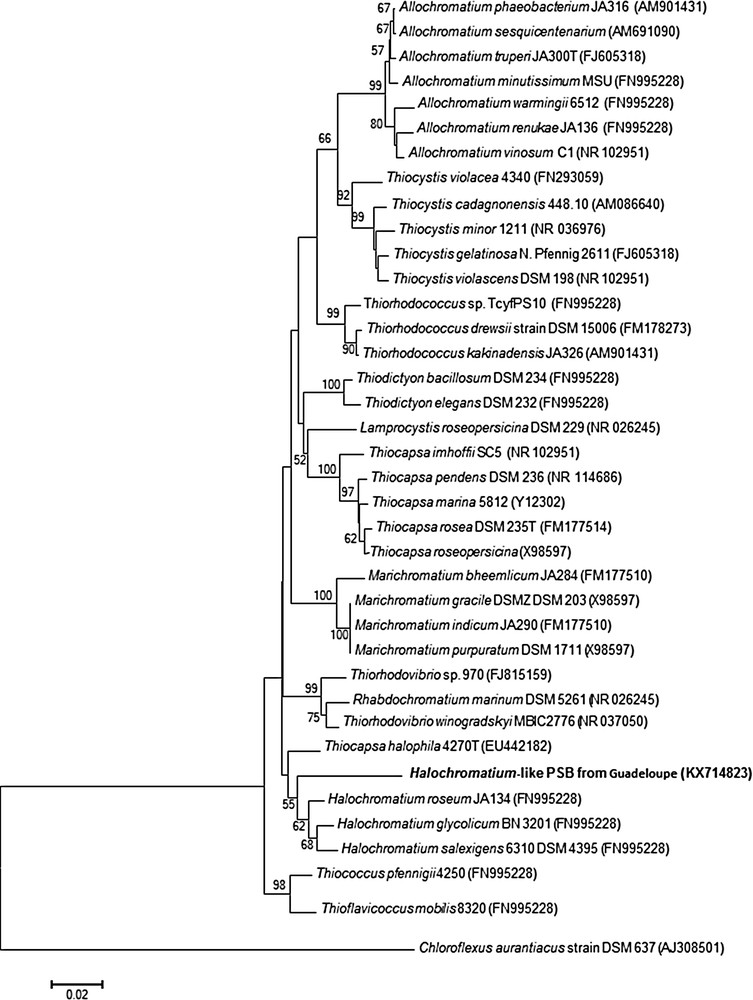
Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences (773 bp) within related species of the family Chromatiaceae. The phylogenetic tree was constructed using the Clustal X [28] and MEGA version 5.2 [29] programs. The bootstrap values obtained by 1000 replications [31] over 50% are beside nodes. The outgroup was specified as strain Chloroflexus aurantiacus DSM 637 gi|15551748|emb|AJ308501.1|) Bar, 2 nucleotide substitutions per 100 nucleotides.
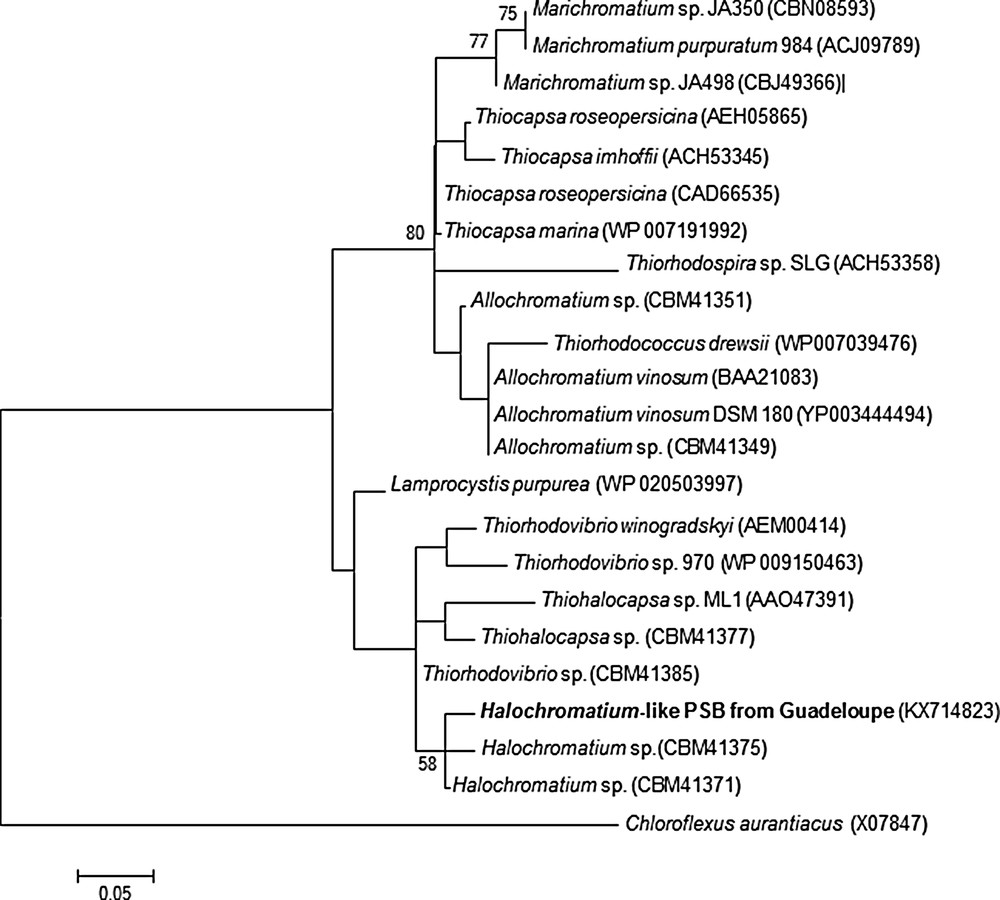
Maximum-likelihood phylogenetic tree based on an amino acid sequence of the PufM products (55 aa) within related species of the family Chromatiaceae. The phylogenetic tree was constructed using the Clustal X [28] and MEGA version 5.2 [29] programs. The bootstrap values obtained by 1000 replications [31] are beside nodes. A green filamentous bacterium, Chloroflexus aurantiacus (gi|40371|emb|X07847.1|), was used as an outgroup. Bar, 5 amino acids substitutions per 100 amino acids.
4 Discussion
Members of Chromatiaceae inhabit anaerobic and sulfide-rich compartments of various aquatic environments from moist and muddy soils to ponds, lakes, rivers, sulfur springs, salt lakes, estuaries, and marine habitats. Major phylogenetic branches within the Chromatiaceae can be distinguished based on their salt requirement. Thus, Imhoff et al. [42] have suggested that salt requirement should be considered for the characterization of “genetically” defined genera, enabling the differentiation between: (i) halophilic species, such as typical marine species like Halochromatium salexigens, Hch. glycolicum, Marichromatium gracile and Mch. purpuratum and (ii) freshwater species, such as Allochromatium vinosum and relatives. A marine species may belong however to a separate genus according to its phylogeny rather than to its affinity for salt.
The environmental PSB novel phenotype described herein represents a marine species evolving in a sulfidic marine environment with a salinity comprised between 3.2 and 3.5%. Halochromatium-like PSB from Guadeloupe individuals are spherical (6.75 μm in diameter), whereas the genera Thiorhodovibrio and Halochromatium, are spiral and rod-shaped, respectively. Furthermore, a phylogenetic analysis based on 16S rDNA sequences and/or puf sequences demonstrated that Halochromatium-like PSB from Guadeloupe is clearly divergent from the described genera (with identity < 93%). In summary, the molecular and phenotypic data support the affiliation of Halochromatium-like PSB from Guadeloupe with various clades within the genus Halochromatium, but not to the genus Thiorhodovibrio. Thus, the strain collected from the marine mangrove environment may correspond to a new species of Halochromatium.
The sulfur metabolism of this new species was confirmed by EDX analysis. The use of ESEM to observe samples that have not yet been dehydrated in solvents (such as ethanol or acetone), coupled with elemental sulfur detection by EDXS analysis, is not common in microbiological investigations. We used these two techniques to assess the ability of this new environmental phenotype of Halochromatium to store elemental sulfur within its cytoplasm. X-ray absorption spectroscopy has already been used to study many thioautotrophic bacteria, including photosynthetic sulfur bacteria, to demonstrate that sulfur stored in the cytoplasm of these organisms is the solid S8 form [43]. We showed that Halochromatium-like PSB from Guadeloupe cells store elemental sulfur in their cytoplasm when exposed to sulfides from their environment.
In marine coastal ecosystems, microbial mats are usually composed of various prokaryotes (cyanobacteria, thioautotrophic bacteria, heterotrophic bacteria, archae) as well as of diverse eukaryotic microalgae, such as benthic diatoms. These organisms are usually organized according to their physiology into vertical laminated structures with the top layer colonized by cyanobacteria/diatoms or, in a few cases, colorless sulfur bacteria belonging to Beggiatoa [10]. In the marine mangrove of Guadeloupe, filamentous cyanobacterial mats [35] and colorless filamentous Beggiatoa mats [4] have been recently described. These micro-organisms usually all live in the same compartment and are most often mixed together (Fig. 1a). The sediments associated with these microbial mats contain large amounts of clay and have high TOC and TN levels and high TOC/TN ratios, with low total carbonate content and δ13C levels [21]. These findings are consistent with the presence of sulfur-oxidizing microbial mats (with gamma and epsilon proteobacteria [4,44]) close to the roots of mangrove trees. The classical organization of bacterial communities in well-delimited, successive layers is less clear in marine mangroves, because of the “fluidity” of the sediment. Thus, bacteria can move easily from deeper layers to the top layer according to environmental factors (O2, pH, organic matter, etc.). This may explain the existence of temporary blooms (no more than a few days) at the surface of mangrove sediments of Halochromatium-like PSB from Guadeloupe (Fig. 1.a). It was previously suggested that mass blooms of anoxygenic phototrophic bacteria can develop at the sediment surface if the organic matter input is sufficiently high [9]. Recently, Gontharet et al. [22] showed that the grain size distribution of sediment particles and total inorganic content are the main parameters that influence the occurrence of microbial mats at the sediment–water interface in marine mangroves of Guadeloupe. However, seasonal changes in nutrients (such as nitrogen contents, organic matter, etc.) are unknown for mangrove sediments in Guadeloupe.
The biodiversity, ecology, and physiology of anoxygenic purple bacterial mats is less well documented than for oxygenic phototrophic organisms, such as cyanobacteria and/or micro-algae, such as diatoms. According to Hubas et al. [10], sediment cohesion in Roscoff Aber Bay was enhanced and sediment was stabilized because of the abundance of PSB. High amounts of extracellular polymeric substances (EPS) produced by microbial mats may glue the sediment grains together, enhancing the stability of the sediment against erosion in tidal environments [45]. However, Caribbean mangroves are submitted to low tidal ranges (maximum 40 cm), especially in “La Manche à Eau” [46], and marine microbial mats never break the surface of the water. Halochromatium-like PSB from Guadeloupe mats consist of a very large number of rapidly moving independent cells. Thus, these mats are poor in exopolymeric substances and could not be involved in the stabilization of the sediment as they are easily dispersed by low water currents in the water column produced by fish for example (personal observations).
Anoxygenic phototrophic bacteria play an important role in the biogeochemical cycling of carbon and sulfur in stratified ecosystems [47]. Thus, they probably also play a key role in carbon and sulfur turnover in mangrove sediments. The contribution of anoxygenic marine mangrove bacterial populations to C and N cycles and their role in sustaining local food webs are underestimated. The true contribution of PSB communities to total primary production in marine mangroves is poorly understood and requires further study.


