1 Introduction
1.1 Functions of the cellular prion protein (PrPC)
Transmissible spongiform encaphalopathies (TSEs) are fatal neurodegenerative diseases that are characterized by the accumulation of a protease resistant form (PrPSC) of a cellular protein (PrPC), that is normally sensitive to digestion 〚1〛. There has been considerable speculation as to the function of the cellular prion protein (PrPC), which is a widely expressed glycoprotein, anchored in the plasma membrane via a glycosylphosphatidylinosol link. PrPC null mice appear to develop and reproduce normally and are no longer susceptible to TSEs, suggesting that normally PrPC has no essential function that cannot be compensated 〚2〛. However evidence suggests that PrPC performs a multiplicity of functions including the binding of Cu2+ which may be associated with superoxide dismutase activity 〚3〛; the capacity to bind cell surface receptors, in particular that for laminin 〚4〛; the transduction of extracellular signals impinging at the plasma membrane 〚5〛; and the ability to bind nucleic acids 〚6–8〛.
The long standing interest of our laboratory with respect to nucleic acid binding proteins, and in particular the nucleic acid chaperone functions of retroviral nucleocapsid proteins (NC), prompted us to examine a possible activity of PrPC in the chaperoning of viral nucleic acids in comparison with the well established, specific chaperone activity of the HIV-1 nucleocapsid protein NCP7 〚9–10〛.
1.2 Nucleic acid chaperone functions of NCP7
The nucleic acid chaperone properties of NCP7 with respect to retroviral replication have been reviewed elsewhere 〚9–10〛 and are summarized in figure 1. In general, nucleic acid chaperone activity depends on the capacity of the chaperone protein to bind to a nucleic acid and destabilise its secondary structure. Protein–nucleic acid and protein–protein interactions destabilize nucleic acid structures of intermediate stability. This results in the progressive accumulation of the elements in their most stable structural conformation. In this way the activity of a nucleic acid chaperone can for example promote the annealing of complementary strands of nucleic acids under physiological conditions when in the absence of chaperones high temperatures and salt concentrations would be required to break down the single-stranded initial structure to allow hybridization. However because chaperone activity destabilizes structures of intermediate stability, only stable interactions between appropriate complementary partners will be facilitated.
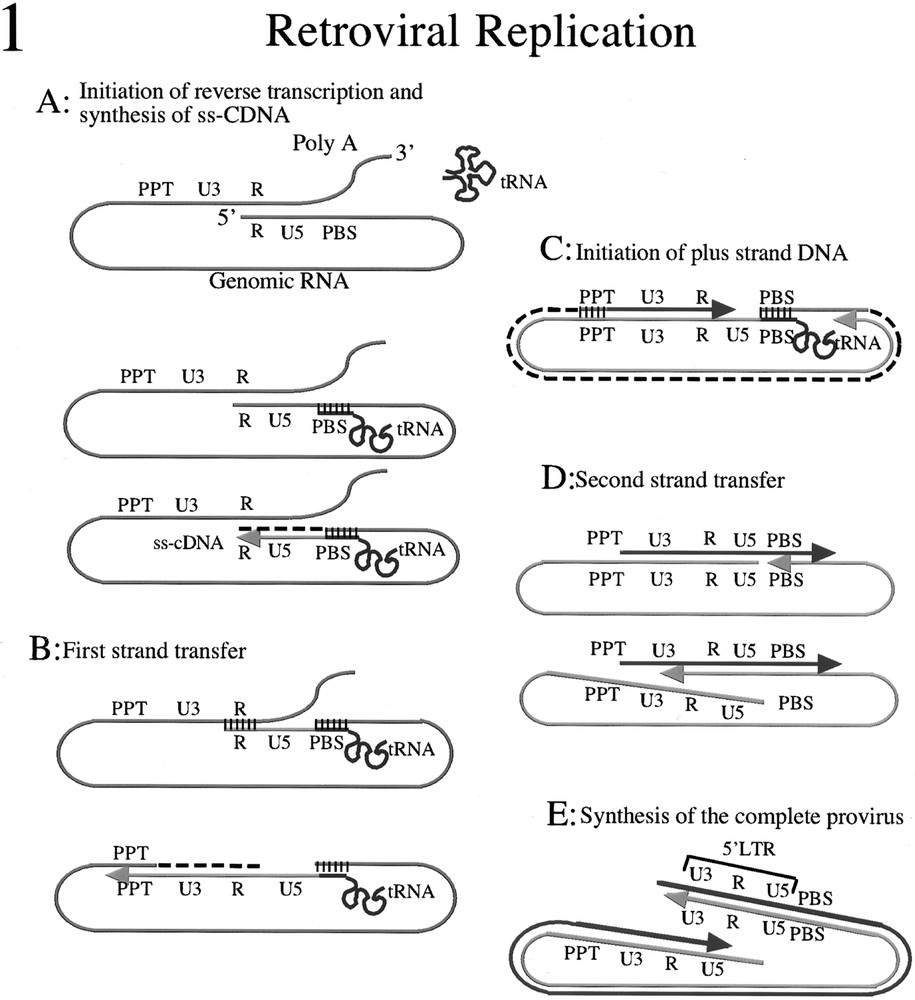
NCP7 nucleic acid chaperone functions during retroviral replication. A. Initiation of reverse transcription requires the annealing of the tRNALys (red line) to the primer binding site on genomic RNA (green line), reverse transcriptase and NCP7. NCP7 functions to destabilize the secondary structure of the nucleic acids and allow hybridization of the complementary sequences in the tRNA and the PBS. Elongation of the primer then occurs towards the 5ˈ terminal of the genomic RNA to produce the ss-cDNA (blue line) and the genomic RNA template is degraded by the RNAse H activity of RT (dashed black line). Hybridization of complementary sequences is indicated by the group of short parallel black lines. B. First-strand transfer is required for the synthesis of the 3ˈ LTR. The complementary R sequences of the ss-cDNA and the 3ˈ genomic RNA must hybridize to allow the continued elongation of the minus strand cDNA (blue line) using the genomic RNA as template (green line). The hybridization of these complementary sequences under physiological conditions requires the nucleic acid chaperone activity of NCP7. Elongation of the cDNA continues towards the PBS and RNAse H activity degrades the genomic RNA (dashed black line) with the exception of a 16 nucleotide oligomer designated the polypurine tract (PPT) which is resistant to RNAse H activity. Hybridization of complementary sequences is indicated by the group of short vertical black lines. C. The PPT serves as the initiation site of the plus strand DNA (pink line) with 5ˈ LTR. The genomic RNA template is degraded (dashed black line) by the RNAse H activity of RT as minus strand cDNA synthesis (blue line) progresses towards the PBS. D. The second strand transfer occurs when RT displaces the PBS and copies the part of tRNA which is complementary to the PBS. The complementary sequences of the PBS in the plus and minus strand DNA can then anneal, a process which requires the nucleic acid chaperone activity of NCP7 under physiological conditions. E. The synthesis of the complete provirus can then occur by elongation of the plus strand cDNA (pink line) using the minus strand cDNA (blue line) as template.
2 Results
2.1 PrP mimics NCP7 chaperone activity in tRNALys annealing and initiation of ss-cDNA synthesis by RT
The nucleic acid chaperone activity of NCP7 is essential during several phases of the life cycle of HIV-1. After receptor binding, penetration of the viral core inside a cell and uncoating, the chaperone activity is required for the generation of the proviral DNA from the genomic RNA template. The initial step in a sequence of events is the specific production of the HIV strong stop cDNA (ss-cDNA) from the RNA template and requires the annealing of a tRNALys primer to the complementary primer binding site (PBS) in the 5’ region of the genomic RNA and reverse transcriptase (RT) to catalyse elongation of the ss-cDNA. That a nucleic acid chaperone is also required for this process is illustrated by the failure of the reaction to proceed under physiological conditions in vitro in the absence of NCP7 (figure 2, lane 1 and 5) 〚10〛. However, in the presence of NCP7 annealing of the radioactively labelled tRNALys occurs allowing production of the ss-cDNA by RT (figure 2, lanes 2–4). The capacity to chaperone the annealing of the tRNA to the PBS is a highly specific property of retroviral NC proteins that is not shared by well characterized cellular nucleic acid chaperones such as Rec A, p53 and others 〚10, 11〛. For this reason we were very surprised to find that PrPC of human origin could substitute for NCP7 in this reaction (figure 2, lanes 6–8).
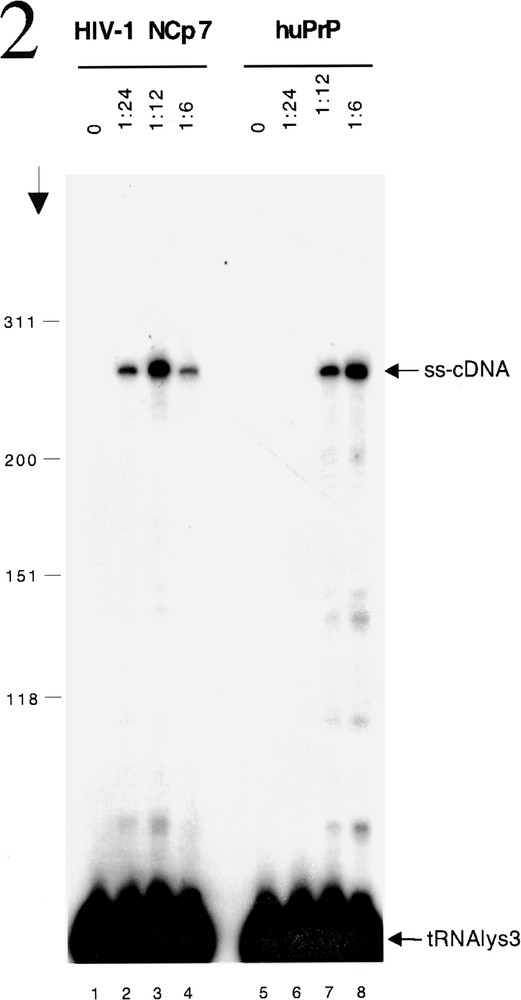
HuPrP chaperones tRNALys annealing and initiation of ss-cDNA production by RT. HIV-1 5’ RNA and 32P-labelled tRNALys were incubated with or without NCP7 or huPrP at the protein to nucleic acid ratios indicated above the lanes. HIV-1 RT p66-p51 was added together with dNTPs to allow initiation of reverse transcription. Arrow shows the direction of electrophoresis and markers (in nucleotides) are shown on the left. ss-cDNA and tRNALys are indicated on the right. The formation of ss-cDNA requires the binding of the tRNALys to the primer binding and site and this does not occur in the absence of an appropriate nucleic acid chaperone (lanes 1 and 5). As expected NCP7 is able to chaperone this process which is required for the initiation of reverse transcription (lanes 2–4) as shown by the presence of the radioactive tRNALys primer present in the ss-cDNA formed by the extension of the primer bound to the genomic RNA template. Suprisingly huPRP shows a very similar activity (lanes 6–8).
2.2 PrP mimics NCP7 during strand transfers required to generate LTRs of HIV-1 proviral DNA
In order to generate the proviral DNA flanked by the LTRs the ss-cDNA must be transfered to the 3’ end of the genomic RNA to act as primer for complete minus strand DNA (cDNA(-)) synthesis. For this process to occur RT with RNAse H activity, the 5’ and 3’ RNA sequences each containing the R sequences present in these regions of the genomic RNA and the nucleic acid chaperone activity of a retroviral NC are needed 〚9〛. In vitro this can be modelled using a labelled oligonucleotide complementary to the primer binding site as a primer together with the other elements and NC. In the absence of NC no specific reaction products are visible (figure 3, lane 1). However in the presence of NCP7 a dose-dependent synthesis of the specific ss-cDNA and HIV-1 cDNA(-) reaction products occurs (figure 3, lanes 2-4). HuPrP can replace NCP7 at similar concentrations in this assay of strand transfer (figure 3, lanes 6–8) indicating that huPrP has a chaperoning activity similar to NCP7. In vitro experiments also established that huPrP can replace NCP7 during the second strand transfer (data not shown) 〚12〛.

HuPrP chaperones the minus strand DNA transfer required for HIV-1 proviral DNA synthesis. The in vitro model of the HIV-1 strand transfer process uses template 5’ and 3’ RNAs each containing the R sequence and corresponding to the 5’ and 3’ ends of the HIV-1 genomic RNA, and a 32P-labelled DNA complementary to the primer binding site. The reaction also requires RT with RNAseH activity and an appropriate nucleic acid chaperone. Following synthesis of ss-cDNA the 5’ RNA template is degraded by the RT-RNAse H activity to allow transfer of the ss-cDNA to the 3’ RNA. The annealing of the ss-cDNA requires an appropriate nucleic acid chaperone to allow the ss-cDNA to act as primer for consecutive steps of reverse transcription during the process of proviral DNA synthesis. In the absence of chaperone the strand transfer does not occur. NCP7 (lanes 2–4) or huPrP (lanes 5–8), in the protein to nucleic acid molar ratios indicated above the corresponding lanes, are able to chaperone the strand transfer as indicated by the formation of the expected specific reaction product. Arrow indicates the direction of electrophoresis, molecular weights (in nucleotides) are indicated on the left and the positions of ss-cDNA and the reaction product (cDNA(-)) are indicated on the right.
2.3 PrP mimics NCP7 with respect to the inhibition of self-primed reverse transcription
Besides chaperoning the specific interaction between complementary nucleic acid sequences (primer binding, strand transfers), that must occur for the synthesis of proviral DNA from the genomic RNA template, NC proteins also function to inhibit non-specific replicative events that can take place. The repair of nicks in the genome or refolding of the RNA template can cause self-initiation of reverse transcription. Needless to say these non-specific phenomena will prevent the faithful production of proviral DNA. Retroviral NC proteins bind with low affinity over the whole length of the RNA template. The chaperone activity, which destabilizes RNA structures of intermediate stability, thus prevents self initiation 〚9〛. In the absence of a nucleic acid chaperone non-specific cDNA is synthesized as a result of self-priming of the RNA template in the presence of RT (figure 4, lanes 1 and 5). However, when NCP7 (figure 4, lanes 2-4) or huPrPC (figure 4, lanes 6–8) is included in the reaction, self priming is inhibited in a dose dependent manner.
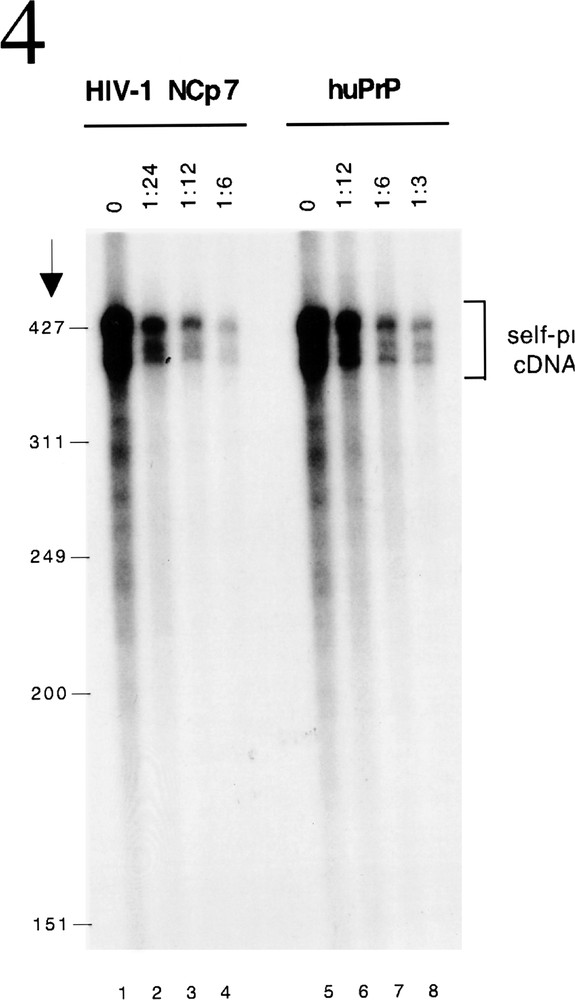
HuPrP inhibits self priming of reverse transcription. Self priming occurs when RNA fold and hybridization of moderate structural stability occurs between short complementary sequences on the same RNA to prime reverse transcription. This can be assayed in vitro using HIV-1 3’ RNA as template, 32P-labelled dCTP and RT (lanes 1 and 5). The nucleic acid chaperoning activity of NCP7 (lanes 2–4) or huPrPC (lanese 6–8) inhibits this process in a dose-dependent manner.
2.4 PrP mimics NCP7 with respect to the dimerization of HIV-1 genomic RNA
In viral particles the HIV-1 retroviral genome is present as a dimer coated with about 2 000 molecules of NCP7 with the replication primer tRNALys annealed to the primer binding site. The dimerization of genomic RNA also functions to form the viral nucleocapsid and participates in the process of viral particle budding 〚9〛. The dimerization process can be investigated in vitro using 5ˈ leader RNA, labelled tRNALys and nucleic acid chaperones. In the absence of an appropriate chaperone there is neither primer binding nor dimerization (figure 5A and B, lanes 1 and 5). However in the presence of NCP7 binding of the tRNALys occurs and the dose-dependent formation of RNA dimers (and in vitro oligomers) can be observed (figure 5A and B, lanes 2–4). It is very suprising that huPrPC shows an identical activity (figure 5A and B, lanes 6–8). It is well established that this is a specific characteristic of retroviral NC proteins and similar activity is not observed with other well characterized nucleic acid binding and chaperone proteins of either viral, bacterial or cellular origin e.g. RT, HIV-1 Vpr and Integrase, T4 gp32, RecA and p53 〚9–11〛.
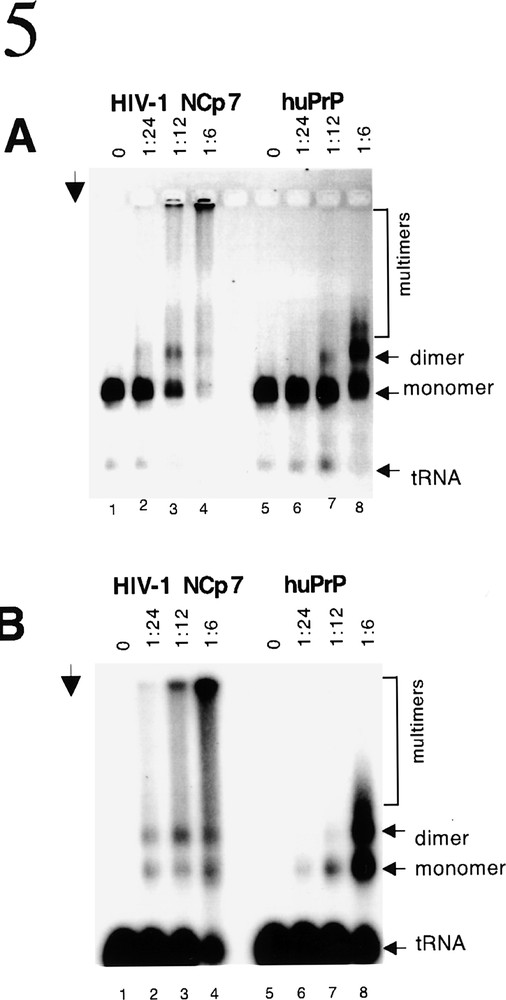
PrP and NCP7 dimerize HIV-1 genomic RNA. In viral particles HIV-1 genomic RNA is dimeric and primer tRNALys is annealed to the primer binding site. This process, which is necessary for formation of nucleocapsid and the budding of viral particles, requires an appropriate nucleic acid chaperone. HIV-1 5’ RNA was incubated with 32P-labelled primer tRNALys under physiological conditions. A and B show the same gel analysed by ethidium bromide staining of nucleic acids (A) and autoradiography (B) to visualize the annealing of the 32P-labelled tRNALys. In the absence of an appropriate nucleic acid chaperone there is no annealing of the tRNALys to the PBS located on the HIV-1 5’ genomic RNA and no dimerization (lanes 1 and 5). Both NCP7 and huPrP are able to chaperone these reactions as indicated by the presence of radioactive tRNALys annealed to monomers, dimers and (because the reaction is being carried out in vitro) multimers of the 5’ genomic RNA (lanes 2–4 and lanes 6–8). Arrow indicates the direction of electrophoresis. The positions of monomers, dimers and multimers of the genomic RNA are indicated on the right. Molar ratios of protein to nucleic acids are indicated above each lane.
3 Discussion
The finding that huPrPC behaves as a nucleic acid chaperone confirms and extends previous evidence that PrPC is a nucleic acid binding protein 〚6–8〛. The nucleic acid binding activity can be demonstrated using genomic RNA of HIV-1 by gel shift assay 〚12〛. Interestingly PrP also interacts with viral DNA to form large nucleoprotein complexes similar to those formed by NC binding to DNA as revealed by dark field electron microscopy 〚12〛. Indeed it was these findings that prompted us to further investigate a possible function for PrPC as a nucleic acid chaperone 〚12, 13〛. Our results, which show clearly that huPrP is a molecular mimic of NCP7, and retroviral NC in general, are of particularly great interest. HuPrPC has all the specific chaperone functions of HIV-1 NCP7 being able to chaperone tRNALys annealing to primer binding site, both the minus and plus strand transfers required for synthesis of proviral DNA flanked by LTR and dimerization of genomic RNA. Furthermore huPrP also possesses the capacity to inhibit self-initiation during elongation by RT to ensure the fidelity of the reverse transcription process. Many cellular or indeed viral proteins have been identified with nucleic acid chaperone properties, but apart from NC proteins, no other has these functions 〚9, 10〛. These nucleic acid chaperone properties are not unique to huPrPC because similar results were obtained with PrPC of ovine 〚12, 13〛 and murine origin. Studies of the nucleic acid chaperone domain of huPrPC show that the nucleic acid chaperone properties map to the unstructured N-terminal region 〚12, 13〛. Interestingly there is no evident sequence or structural homology with identified retroviral NC proteins.
One question is: why should a cellular protein have these functions? The nucleic acid chaperone functions that PrPC possesses are not apparently essential for any identified cellular function, and such chaperone activity might be expected to facilitate infection by retroviruses. On the contrary, a hypothesis under investigation in our laboratory is that PrPC may function as a defence mechanism against retroviral infection. There is some preliminary evidence to support this. In the central nervous system, where cellular levels of expression of PrPC are highest, astrocytes are the major neural cells infected by HIV-1. Part of the cellular response of astrocytes to HIV-1 infection is to increase expression of the PrPC mRNA 〚14〛. Astrocytes infected with HIV-1 do not normally produce infective particles 〚14〛. We have compared the levels of expression of PrPC in established stable MLV-based helper cell lines and in the parent cell lines from which they were derived. So far we have always found that the expression of PrPC is either reduced or indeed not detectable in the helper cell lines although PrPC is readily detectable in the parent cell line (E.D. unpublished). This would suggest that high levels of expression of PrPC may not be compatible with efficient production of retrovirus (i.e. HIV, MLV).
Preliminary evidence suggests that PrPC interacts with the GAG polyprotein of HIV-1 from which NCP7 is formed by proteolytic maturation. PrP might compete with NCP7 for binding sites on the HIV-1 genomic RNA and thus perturb discrete phases of the viral life cycle that we have yet to identify. GAG plays a critical function in the assembly of viral particles, perhaps PrP–GAG interactions may function to inhibit assembly. If PrPSC were to remain bound to genomic RNA in the viral core, the question would arise as to whether viruses might behave as vectors for PrPsc. If this were the case one might expect transmissible spongiform encaphalopathies to be more rapidly disseminated and produce lesions more quickly in animals co-infected with PrPSC and retroviruses. Indeed this has been reported in the mouse-MLV system 〚15〛.
On the other hand interactions between PrP and viral nucleic acids might activate secondary messenger cascades that may influence processes required for efficient viral replication or perhaps induce apoptosis of the host cell. This would explain the absence, or the reduced expression of PrPC in helper cells selected for efficient stable virus production. However, despite the fact that PrPC has been implicated in signal transduction 〚5〛, the existence of such a mechanism remains to be established. Clearly the elucidation of such a phenomenon may be of great interest with respect to the development of new anti-viral strategies.
Acknowledgements
This research has been generously supported by ANRS, Sidaction and the MGEN. ED is beneficiary of a fellowship from the ANRS.



