1 Introduction
The dynamic process of signal transduction involves the concerted action of both protein kinases and protein phosphatases. Serine/threonine phosphorylation is crucial event for regulating cellular processes, since more than 98% of protein phosphorylation occurs on serine and threonine residues. PP1 proteins are evolutionarily conserved enzymes that represent a substantial fraction of serine/threonine phosphatase activity. In cells, PP1 activity always results from the interaction of a PP1c with a variety of targeting or interacting proteins that generate a family of PP1 holoenzymes and determine substrate selectivity [1,2]. Consequently, identification of new PP1 partners corresponding to new holoenzymes is important for future in vivo control of protein phosphorylation. The number of known PP1c interacting proteins is continuously increasing and to date, more than 50 unique eukaryotic proteins have been already identified (for complete information see our website). Biochemical, structural and X-ray crystallography analysis of PP1 interacting proteins established that a short amino acid R/K-x(0,1)-V-x-F peptide sequence represents a widespread consensus motif for recognition and binding of distinct regulatory subunits to PP1c [3,4]. We have recently demonstrated that the anti-apoptotic protein Bcl-2 is also a new PP1 interacting protein that binds PP1c (through the R-I-V-A-F sequence analogous to the consensus motif R/K-x(0,1)-V-x-F and targets PP1c to its substrate Bad [5,6].
This report established that the simultaneous presence of two distinct consensus PP1c docking motifs R/K-x(0,1)-V-x-F and F-x-x-R/K-x-R/K may be used as a signature to identify new putatively interacting PP1c proteins. To allow easy and rapid identification of new potential PP1 interacting target proteins, we have created a new bioinformatic tool, represented by the website http://pp1signature.pasteur.fr/.
2 Materials and methods
2.1 Immunoprecipitation and western blotting analysis
IL-4-stimulated TS1αβ cells (1×107) were lysed for 20 min at 4 °C in lysis buffer (50 mM Tris HCl pH 8, 1% NP-40, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 10% glycerol and protease inhibitor cocktail). Lysates were immunoprecipitated with the appropriate antibody and protein A-Sepharose was added for 1 h at 4 °C. After washing, immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose and blocked with 5% non-fat dry milk in Tris-buffered saline (TBS, 20 mM Tris HCl pH 7.5, 150 mM NaCl) and incubated with primary antibody in TBS/0.5% non-fat dry milk. The membrane was washed with 0.05% Tween-20 in TBS and incubated with PO-conjugated secondary antibody. After washing, proteins were developed using the ECL system.
Anti-PP1α, anti-p85 PI3-K, anti-p110 PI3-K, anti-Hsp70, anti-Bcl-2, anti-Ras and anti-CD4 antibodies were from UBI (Lake Placid, NY), Calbiochem (La Jolla, CA) or Transduction Laboratories (Lexington, KY).
2.2 Peptide synthesis and protein–protein interaction competition
Peptides from Bcl-xL and Bad were prepared as described [7,8] by automated spot synthesis (Abimed, Langerfeld, Germany) onto an amino-derivatized cellulose membrane, immobilized by their C-termini via a polyethylene glycol spacer and N-terminal acetylated.
The interaction of the targeting subunits with PP1 was competed by the F-x-x-R-x-R, (GDEFELRYRRAF) and R-I-V-A-F (NWGRIVAFFSF) peptides. Lysates were immunoprecipitated with anti-PP1α antibody and protein A Sepharose was added. The interaction was competed by incubation with the peptides (30 min, room temperature). After washing, immunoprecipitates were transferred to nitrocellulose and blotted with the corresponding antibody.
3 Results and discussion
3.1 F-x-x-R/K-x-R/K: a new PP1 binding motif in Bcl-xL and Bcl-w proteins
Using a murine T cell line that can be propagated independently in the presence of IL-2 IL-4 or IL-9 [9], we have recently described that PP1α is a Ras-activated phosphatase that dephosphorylates Bad, a pro-apoptotic member of the Bcl-2 family proteins, prior to induce apoptosis in response to IL-2 deprivation [5]. As shown in Table 1, two sequences (R-I-V-A-F or R-L-V-A-F) analogous to the R/K-x(0,1)-V-x-F PP1 binding consensus motif are present in the BH1 domain of three Bcl-2 family proteins. We have shown that Bcl-2, Bcl-xL and Bcl-w require these motifs for binding and targeting PP1α to Bad. Based on biochemical competition studies, we demonstrate that Bcl-2 is a new PP1 interacting protein that binds PP1α through the R-(IL)-V-A-F sequence and targets the phosphatase to Bad [5]. In addition to this canonical docking consensus site, we have also characterized a new PP1c docking site in Bcl-xL and Bcl-w [10]. The new docking site comprises a motif analogous to the 140-F-E-M-R-R-K-146 sequence identified as a PP1c binding site for the small cytosolic protein inhibitor-2 (I-2) (Fig. 1A) [11,12]. A new F-x-x-R/K-x-R/K consensus sequence similar to PP1α docking sites in I-2, Bcl-xL and Bcl-w is also present in known PP1c binding proteins (Fig. 1A). Mutation of the critical F and R residues strongly reduces binding of Bcl-xL to PP1c (Fig. 1B).
A canonical R/K-x(0,1)-V-x-F docking motif for PP1c in Bcl-2 proteins
| Bcl-2 | BH1 domains | Residues | Refer- |
| proteins | ences | ||
| 1-Bcl-2 | ELFRDGVNWG RIVAF FEFGG | 143–147 | [6] |
| 2-Bcl-xL | ELFRDGVNWG RIVAF FSFGGAL | 139–143 | [10] |
| 3-Bcl-w | ELFQGGPNWG RLVAF FVFGA | 95–99 | [10] |
| Consensus : R-I/L-VAF |

F-x-x-R/K-x-R/K: a new PP1c docking-site consensus motif in known and putative PP1c-interacting proteins. (A) Sequence alignment of published PP1 interacting proteins in the vicinity of F-x-x-R/K-x-R/K motif. This motif is observed in the anti-apoptotic molecules Bcl-2, Bcl-xL and Bcl-w. (B) PP1c binding assay on cellulose-bound Bcl-xL and Bad peptides. Membrane with Bcl-xL or Bad peptides containing the F-x-x-R/K-x-R/K motif, as well as peptides containing mutated motif were incubated with purified PP1c, followed by anti-PP1α antibody and PO-conjugated secondary antibody. The F-x-x-R/K-x-R/K motif is in bold. Mutated amino acids into the motif are in bold and underlined. Similar results were obtained in three independent experiments.
The new PP1c binding motif is also present in Bad, a proapoptotic member of the Bcl-2 family (Fig. 1B). Mutation of serine residue in the vicinity of the F-xx-R/K-xR/K motif affects binding of Bad to PP1c, although the affinity depends on the type mutation. When the Ser amino acid is replaced by an acidic Asp amino acid, mimicking a phosphorylated amino acid, the binding of the PP1c is inhibited. This result suggests that phosphorylation of Bad on the regulatory Ser-136 while allowing binding of Bad to 14–3–3 [13,14] may also inhibit binding to PP1c (Fig. 1B). Unlike Bcl-xL or Bcl-w, this motif is located outside of the BH3 domain immediately upstream of the regulatory Ser-136.
3.2 Predictive signature for PP1c interactions: simultaneous presence of R/K-x(0,1)-V-x-F and F-x-x-R/K-x-R/K motifs in some characterized PP1 binding proteins
The statistical analysis of sequences in the Swissprot Release 40 indicate that, while 16% of the data library contains one of the motifs, only 3.9% share the two motifs (Table 2). The reduced number of proteins with the two distinct PP1c docking motifs clearly supports the concept of a PP1 predictive signature. In addition, the finding that most characterized PP1-binding proteins share the two motifs sustained this concept and has motivated the Institut Pasteur to create and to maintain the http://PP1signature.pasteur.fr website as a new bioinformatic tool to identify novel putative PP1 interacting proteins. To experimentally validate this concept, we have demonstrated a PP1c association with four candidates, randomly selected in our web site (Fig. 2). We were able to detect association of PP1α to p85 PI3K, p110 PI3K, hsp70 and CD4, four molecules that share both motifs. As a control, in Ras immunoprecipitates we did not observed association of PP1α-Ras, a molecule that contains neither R/K-x(0,1)-V-x-F nor F-x-x-R/K-x-R/K motifs. Interestingly, the association of hsp70 to PP1α is almost undetectable upon competition of PP1α immunoprecipitates with exogenous peptides R and F (for sequence, see materials and methods) corresponding to the two motifs R/K-x(0,1)-V-x-F and F-x-x-R/K-x-R/K, respectively.
Statistical distribution of PP1c docking motifs. Bioinformatic analysis using the Sig program (É. Deveaud, Institut Pasteur) in the Swissprot Release 40 library that contains 101 602 non-redundant protein sequences. For details see general information on the website http://PP1signature.pasteur.fr
| Docking motif | Total of sequences | % of the library |
| R/K-x(0,1)-V-x-F | 16 200 | 16 |
| F-x-x-R/K-x-R/K | 16 273 | 16 |
| R/K-x(0,1)-V-x-F | 4 013 | 3.9 |
| + F-x-x-R/K-x-R/K |
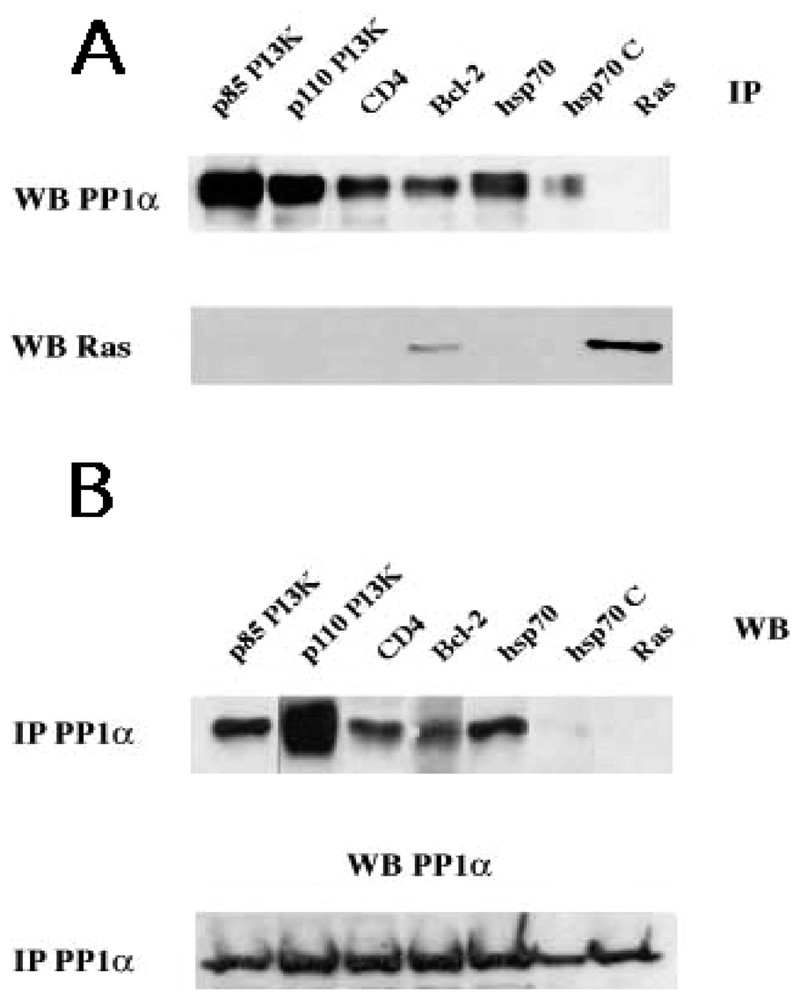
Identification and characterization of new PP1c interacting proteins with a putative signature predicted by sequence analysis. (A) Cytoplasmic lysates from TS1αβ cells were immunoprecipitated with anti-p85 PI3K, anti-p110 PI3K, anti-Hsp70, anti-CD4, anti-Bcl-2 (as a positive control) and anti-Ras (as a negative control) antibodies, transferred to nitrocellulose and immunoblotted with anti-PP1α and anti-Ras. (B) Similarly, cytoplasmic lysates were immunoprecipitated with anti-PP1α, transferred to nitrocellulose and blotted with anti-p85 PI3K, anti-p110 PI3K, and anti-Hsp70, anti-CD4, anti-Bcl-2 and anti-Ras antibodies. For the two panels A and B, the interaction hsp70/PP1α(hsp70C) was competed with 1.5 mM of R (NWGRIVAFFSF) and F (GDEFELRYRRAF) peptides for 30 min at room temperature. Proteins were detected using the ECL system. Similar results were obtained in three independent experiments.
The recent development of penetrating peptides, in addition to the identification of peptide sequences surrounding PP1c docking sites in important regulatory proteins, suggests a new approach for phosphatase-derived drug research. This strategy might be based on the specific ‘peptide knock out’ of an intracellular pathway controlled by a specific PP1 holoenzyme. To develop this academic or therapeutic strategy as well as to facilitate academic research, we propose a new website: http://PP1signature.pasteur.fr, which allows the identification of putative PP1-interacting proteins containing the two distinct PP1c docking consensus motifs represented in the Swissprot library.
Acknowledgements
This work was partially supported by grants from the ‘Association pour la recherche sur le cancer’ (ARC grant 4437), from AVENIR–INSERM and from the Institut Pasteur (grant PTR-60).


