1 Introduction
The root-knot nematode Meloidogyne chitwoodi was first isolated and characterised on potato in northern USA [1]. Since then, this nematode has established itself as a major multi-million dollars pest on potato tubers and is also recorded in western USA, Mexico, Argentina and South Africa [2–6]. More recently, M. chitwoodi was found in the Netherlands, Belgium, Germany and Portugal, where it is considered an emerging pest threatening potato, but also ornamental productions. M. chitwoodi remained unnoticed for long, probably because of populations in relatively low numbers and possible confusion with the prevalent nematode in Northern Europe, Meloidogyne hapla. Economical impact in Europe, through direct loss and downgraded infested tuber stocks, is of concern. In addition, M. chitwoodi is able to develop on other major crops such as alfalfa, wheat, maize, beet, carrot, etc. [7–9]. The risk of spread throughout the European continent is increased by the ability of M. chitwoodi to develop over a large range of temperature. It threatens southern as well as northern Europe, where it can cohabit and be confused with other species of nematodes prevailing in milder climates Meloidogyne arenaria, M. incognita, and M. javanica. A critical issue will therefore be to ensure an early and reliable detection of M. chitwoodi.
Identification of species within the genus Meloidogyne mostly relied on esterase banding patterns in polyacrylamide gel electrophoretic analysis [10–12]. Using this technique, the species M. chitwoodi was characterised by an esterase pattern designated S1 [4]. A second population of nematodes attacking potatoes in Europe was characterised by the absence of an esterase band (null phenotype) and by a different MDH pattern [13]. This biochemical variant was first assigned to M. chitwoodi and was later described as Meloidogyne fallax [14]. Molecular diagnostics aiming at discriminating M. chitwoodi and M. fallax were developed, based on ITS [15,16], IGS [17,18] and SCARs [19].
However a broad view of the genetic diversity of M. chitwoodi and M. fallax is required and more discriminative markers are needed to investigate the genetic diversity at both inter-and intra-specific levels. Different strategies were developed using RFLP [20], RAPD [21] and AFLP markers [22–25] to analyse the genetic diversity of various species of Meloidogyne. They further proved useful to distinguish between as well as within species.
AFLP markers were proved useful to analyse the inter- and intra-specific genetic diversity of various organisms such as plants [26,27], insects [28], fishes [29,30], snakes [31], fungi [32] or bacteria [33]. Different levels of discrimination and analysis were achieved such as identification of interspecific hybrids [30], analysis of patterns of genetic differentiation within an insect species complex [28], phylogeny of rapidly evolving clades [29] or discrimination between closely related species [33]. At the intra-specific level, genetic diversity was assessed and structured into sub-specific taxa [27–29,31,32].
The work presented in this article was aimed at investigating the genetic diversity of M. chitwoodi and M. fallax using AFLP markers. We report here that the group comprising M. chitwoodi and M. fallax separates clearly from the other Meloidogyne species. We also report that M. chitwoodi is characterised by a high genetic diversity and is structured into subgroups that may be undergoing speciation.
2 Material and methods
2.1 Populations of nematodes
Nematode samples originated from farm fields. Codes, specific diagnosis and geographical origins are provided in Table 1. Lines of M. incognita, M. arenaria, M. javanica, M. mayaguensis and M. hapla originate from nematode cultures raised from single egg mass lines, whereas M. chitwoodi and M. fallax correspond to sampled populations. Nematodes were multiplied on susceptible tomato in greenhouses.
Populations of Meloidogyne nematodes used for genetic diversity analysis
| Nematode species | Sample code | Geographic origin |
| M. javanica | 22 | Burkina Faso |
| 23 | Burkina Faso | |
| M. arenaria | 10 | Ivory Coast |
| 28 | French West Indies | |
| 31 | French West Indies | |
| 32 | French West Indies | |
| M. incognita | 9 | Ivory Coast |
| 15 | Thailand | |
| M. mayaguensis | 3 | Ivory Coast |
| 13 | Puerto Rico | |
| M. hapla | 33 | The Netherlands |
| M. chitwoodi | cce | The Netherlands |
| ck | The Netherlands | |
| cu | The Netherlands | |
| cl | The Netherlands | |
| cp | The Netherlands | |
| ca | The Netherlands | |
| cx | The Netherlands | |
| cz | The Netherlands | |
| ccg | Portugal | |
| cch | Argentina | |
| cbh | California | |
| M. fallax | fa | The Netherlands |
| fb | The Netherlands | |
| fh | France |
2.2 Molecular marker analyses
Genomic DNA from lines of M. arenaria, M. javanica, M. incognita, M. mayaguensis and M. hapla was extracted and prepared as described by Fargette et al. [20], and genomic DNA from M. chitwoodi and M. fallax populations was extracted and prepared as described by Zijlstra et al. [16]. DNA samples extracted from M. chitwoodi and M. fallax isolates were checked for contamination as previously described [16,19,34]. The AFLP procedure was conducted under standard conditions as recommended by the manufacturer (AFLP Analysis System I kit, BRL). 150 ng of primary templates were digested by EcoRI and MseI in the reaction buffer provided with the kit for 2 h at 37 °C in a final volume of 25 μl. The mixture was incubated at 70 °C for 15 min to inactivate the restriction endonucleases, put on ice and ligated to EcoRI and MseI adapters with T4 DNA ligase for 2 h at 20 °C in a final volume of 50 μl. A 1:10 dilution of the ligation mixture was performed in TE buffer [35] and 5 μl of diluted mixture was used as template for the non-selective preamplification reaction. Preamplification was performed in the presence of the pre-amp primer mix (containing the preamplification primers) provided by the supplier in a final volume of 51 μl and 1 unit of Taq DNA polymerase for a total of 20 cycles under the following conditions: 94 °C for 30 s, 56 °C for 60 s and 72 °C for 60 s. After completion of the 20 cycles, samples were maintained at 4 °C and preamplification was checked by electrophoresis in 2% agarose gel at 11 v cm−1 and staining with ethidium bromide along with DNA from Arabidopsis thaliana as a control sample and a DNA ladder ranging from 75 bp to 4 kb. Preamplification was considered successful when bands from 100 to 1000 bp were clearly visible. Primers were end-labelled with (γ-33P) ATP as recommended by the supplier (Amersham) and selective amplifications were conducted using a total of seven combinations. The primers contained two selective bases on the EcoRI side and three selective bases on the MseI side. The selective nucleotides of the seven primers were TG/CAA, TC/CTG, AC/CAT, TC/CAA, TC/CAT, AC/CTG, TA/CAA. Selective amplification was conducted as specified by the supplier in a final volume of 20 μl first for 1 cycle at 94 °C for 30 s, 65 °C for 30 s and 72 °C for 60 s, then for 12 cycles, where the annealing temperature is lowered by 0.7 °C at each cycle and finally for a series of 23 cycles at 94 °C for 30 s, 56 °C for 30 s and 72 °C for 60 s. After PCR amplification, samples were analysed by electrophoresis in a 0.4-mm-thick, 5% polyacrylamide gel under denaturing conditions (7.5 M urea). Migration was performed in 0.5 × TBE buffer [35] at 1.4 W cm−1 until bromophenol blue is removed from the gel. The gel was then transferred onto Whatman paper, dried and exposed to autoradiography film (Fuji) for one week.
2.3 Data analysis
Scoring for presence/absence of AFLP bands and data analysis was carried out as described in previous studies [20,21]. These data were analysed using the statistical package Genstat 5 [36] to produce similarity matrices [37]. Dendrograms were constructed according to UPGMA. The topology of the dendrogram shown in Fig. 1 was assessed by a bootstrap test (1000 bootstraps).
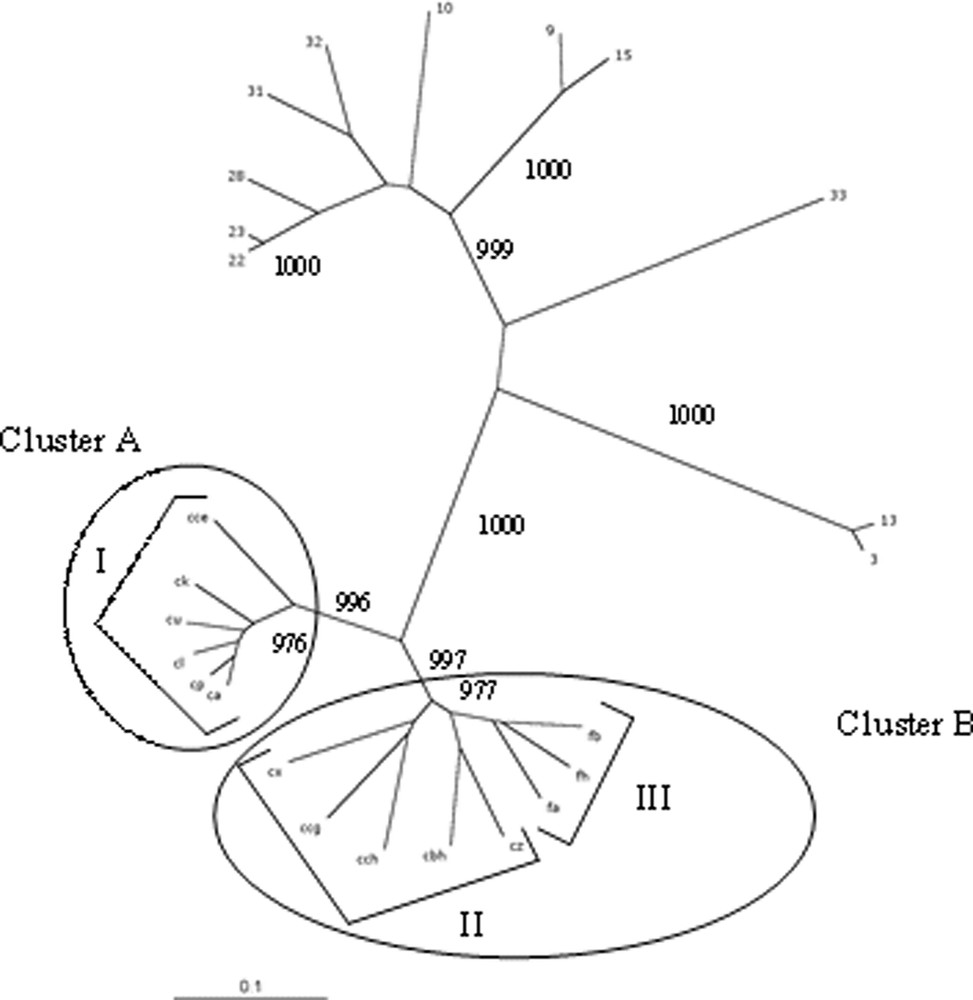
Dendrogram of the relationships between Meloidogyne species. Only bootstrap values higher than 950 are shown.
3 Results
3.1 Genetic relatedness of M. chitwoodi and M. fallax to other Meloidogyne species
The genetic relatedness of M. chitwoodi and M. fallax to other Meloidogyne species was investigated by comparing a total of 14 populations to 11 lines belonging to five species (two lines of M. incognita, four lines of M. arenaria, two lines of M. javanica, one line of M. hapla and two lines of M. mayaguensis). The distribution of the various samples as shown by AFLP analysis is presented in Fig. 1. The distribution shows that the selected representatives of M. incognita, M. arenaria, M. javanica, M. hapla, and M. mayaguensis fall into distinct groups. The M. mayaguensis lines (samples 3 and 13) are clearly separated from the others with a bootstrap value of 1000. Similarly, the M. incognita lines make a separate group characterised by a similar bootstrap value. Although M. arenaria and M. javanica appear clustered, the very high bootstrap value (1000) that supports the M. javanica branch indicates that they are clearly separate groups. M. chitwoodi and M. fallax populations are clustered and clearly separated from the other species of Meloidogyne with a bootstrap value of 1000.
Data from the similarity matrix (Table 2) indicate that the percentage of similarity between the M. chitwoodi/M. fallax group and M. hapla, the M. javanica/M. arenaria group, M. mayaguensis and M. incognita is 49.92%, 37.55%, 46.11% and 42.68%, respectively. Samples 22 and 23 which are M. javanica lines are shown in Fig. 1 to be more closely related to sample 28 (M. arenaria) than the latter is to other M. arenaria representatives (10, 31, and 32). Furthermore, the intraspecific similarity within M. arenaria is lower (79.32%, Table 3) and diversity not as stringently structured (bootstrap values of 849, 637 and 847) than between M. arenaria and M. javanica (82.53% and bootstrap value of 1000).
Genetic relatedness of the M. chitwoodi/M. falllax group to other Meloidogyne species
| Mc/Mf | 77.07 | ||||
| Mh | 49.92 | 100.00 | |||
| Mj/Ma | 37.55 | 55.55 | 82.27 | ||
| Mm | 46.11 | 52.18 | 45.30 | 97.11 | |
| Mi | 42.68 | 53.70 | 71.09 | 48.96 | 92.61 |
| Mc/Mf | Mh | Mj/Ma | Mm | Mi |
Percentage of similarity between groups of Meloidogyne species
| I | 90.11 | |||||||
| II | 69.34 | 81.40 | ||||||
| III | 72.05 | 80.34 | 88.43 | |||||
| Mh | 40.57 | 54.80 | 60.50 | 100.00 | ||||
| Mj | 35.97 | 47.92 | 54.87 | 53.89 | 97.84 | |||
| Ma | 25.42 | 38.76 | 44.04 | 56.38 | 82.53 | 79.32 | ||
| Mm | 37.93 | 50.42 | 55.31 | 52.19 | 54.09 | 40.91 | 97.11 | |
| Mi | 35.14 | 46.46 | 51.48 | 53.70 | 75.92 | 68.68 | 48.96 | 92.61 |
| I | II | III | Mh | Mj | Ma | Mm | Mi |
3.2 Genetic diversity of Meloidogyne chitwoodi and Meloidogyne fallax
Two main and distinct clusters can be identified in Fig. 1. Cluster A comprises the cce, ck, cu, cl, ca, and cp populations of M. chitwoodi (group I). Cluster B is more diverse and comprises populations of M. chitwoodi (group II: cx, ccg, cch, cbh and cz) and M. fallax (group III: fa, fb and fh). Similarity values in Table 4 reflect the presence of these two clusters, characterised by high bootstrap values (998 and 977). With a percentage of similarity of 77% (Table 2), the internal variation within the overall M. chitwoodi/M. fallax group is higher than those of clusters A and B (90.11 and 81.59%, respectively).
Percentage of similarity between clusters of M. chitwoodi and M. fallax
| A | 90.11 | |
| B | 70.36 | 81.59 |
| A | B |
Within group I, the isolate cce branches separately from isolates ck, cu, cl, cp and ca with a bootstrap value of 976 (Fig. 1). Group III is more homogenous (88.43% similarity) than group II (81.4% similarity). Groups I and II (M. chitwoodi) are more distant to each other than group III (M. fallax) is from group II (Table 3).
4 Discussion
The first evidence arising from the data presented here is that outside from the M. chitwoodi/M. fallax group, the observed genetic diversity correlates well with the previously validated species, i.e. M. incognita, M. javanica, M. arenaria, and M. mayaguensis. These species are clearly clustered within groups separated by high bootstrap values (999 or 1000). The only exception to this stringent clustering is the M. hapla line (line 33) with a relatively low value of bootstrap (i.e., 677). This lack of stringency might be a consequence of having only one population of M. hapla in the study, making impossible to assess the intraspecific variability.
The main branch comprising M. chitwoodi and M. fallax is clearly separated from the other Meloidogyne species (large distance and high bootstrap values). The clear separation observed between M. chitwoodi and M. hapla is in agreement with the theory of an early separation of these two sympatric species considered to have occurred between 35 and 43 million years ago [38]. The high level of heterogeneity observed within M. chitwoodi (groups I and II) also confirms the concept of M. chitwoodi being an ancient organism as shown by a high diversity of the mitochondrial DNA [39] and of the AluI satellite DNA [40].
The genetic diversity revealed by AFLP markers within the M. chitwoodi/M. fallax branch indicates the presence of well-structured populations. The main M. chitwoodi/M. fallax group cannot be considered a single species since M. chitwoodi and M. fallax have been shown to be reproductively isolated [41–43]. The minimal clustering must then be considered at the separation of M. fallax samples (group III) from the M. chitwoodi isolates, which is characterised by a bootstrap value of 977. In contrast to the homogeneity of the M. fallax group, M. chitwoodi displays a large heterogeneity with isolates ranging into two different clusters separated by high bootstrap values, i.e. groups I and II. Unlike interspecific markers, AFLP are specifically useful for investigating intraspecific variations and relatedness between closely related entities. Therefore, similarity matrices allow an insight on the relatedness between the different groups through a comparative way. In the present study, we observed that M. fallax is more similar to one subset of M. chitwoodi (group II) than this same subset is to the group I of M. chitwoodi. This observation is in agreement with the analysis of M. fallax satellite DNA, which was found to be more closely related to the two members of the pMcCo2 subfamily of M. chitwoodi satellite DNA than pMcCo2 was to the pMcCo1 subfamily of M. chitwoodi satellite DNA [44]. Although, unlike satellite DNA, AFLP data are not meant for phylogenetic analysis, this organisation of M. chitwoodi into distinct clusters may represent speciation events under process. However, in contrast to M. fallax, it is not possible to define their degree of achievement. Further crossing experiments will be needed to investigate the existence of reproductive isolation between the M. chitwoodi clusters if any. Such a distribution may reflect the facultative parthenogenetic reproductive behaviour of M. chitwoodi as well as the poor ability of these organisms to move and mix and would be compatible with the concept of complex of species. Owing to this low mobility, low encounter probability and trends to parthenogenesis, these populations could be considered as being ‘pseudoclonal’. Furthermore, beside amphimixy, facultative parthenogenesis most likely has a non negligible share in the reproductive process, as shown by the low number of males in M. chitwoodi populations [41].
The observed variation in the width of the genetic basis between groups I and II could also reflect the coexistence of populations characterised by different modes of reproduction. Indeed, biodiversity within amphimictic populations would be displayed differently than in parthenogenetic ones. Under such a hypothesis, these two groups may correlate with different modes of reproduction, hence displaying different degrees of heterogeneity. On the other hand, one cannot exclude either a structuring effect of the host plants on which populations were sampled. However, appropriately designed experiments will be needed to investigate these various hypotheses.
M. fallax is considered to have achieved speciation from at least one population of M. chitwoodi [44], displaying thus a narrower genetic basis as shown in the similarity matrices presented in Table 3. According to the analysis of the AluI satellite DNA from M. fallax, which was found to be less variable than that of M. chitwoodi, this speciation was described as a recent event [44]. The structure of the M. chitwoodi populations could be compatible with a paraphyletic organisation. However, further studies using phylogenetic markers, i.e. DNA sequences, must be conducted to unravel this evolutionary aspect. Associating these phylogenetic analyses to interfertility tests would definitely demonstrate whether the high genetic diversity observed within M. chitwoodi is indeed an image of populations currently undergoing speciation or recently separated as species, a status which could ultimately lead to a revision of the nomenclature, as done with M. fallax.
Acknowledgments
This work was supported by a grant from the E.C. programme FAIR-CT95-0896. The authors are very grateful to Dr C. Zijlstra (Plant Research International B.V., Wageningen, The Netherlands) for providing M. chitwoodi and M. fallax purified DNA.


