1 Introduction
Root-knot nematodes (RKN; Meloidogyne spp.) are economically important plant pathogens in many countries. Like other soil-borne plant pathogens, damage caused by RKN is underestimated because the aboveground symptoms due to RKN infection are often incorrectly attributed to other causal agents, such as bacterial or fungal diseases, nutrient deficiency or drought stress [1]. RKNs display different reproduction modes. Few RKNs can reproduce by the classical amphimixis (e.g., M. carolinensis), while some reproduce by the combination of cross-fertilization and facultative meiotic parthenogenesis (e.g., M. graminicola) and others by obligatory mitotic parthenogenesis (e.g., M. incognita) [2,3]. Sexual RKNs display a narrow host range while parthenogenetic RKNs show a wide host range [3]. This suggested that the reproductive mode influences the pathogenicity level of RKNs. The Meloidogyne genus therefore appears as a model system to investigate the impact of each reproduction system on the maintenance of genetic diversity and intraspecific variability in correlation to the pathogenicity [3]. For instance, M. chitwoodi, a meiotic parthenogenetic nematode, has a wide host range and has shown to have significant intraspecific variability and genetic structure between four populations from the USA [4]. In addition, several obligatory mitotic parthenogenetic nematodes having a larger host range were studied (e.g., M. enterolobii), and were characterized by a genetically homogeneous organization [5]. More exhaustive data then need to be generated to explore the potential contribution of each reproductive mode in the genetic structure organization and intraspecific variability.
Meloidogyne graminicola[6] – a facultative meiotic parthenogenetic species – is the most common RKN infecting Asian rice (Oryza sativa L.) and this species can cause significant yield losses in all rice agro-ecosystems in Asia [7–11]. In upland conditions and shallow intermittently flooded land, M. graminicola is considered to be the most damaging Meloidogyne species on rice [1]. These mostly rain-fed rice agro-ecosystems occupy about one-third of the rice-growing area in Asia [12], indicating that there is a need for new rice varieties resistant or tolerant to M. graminicola [13]. Nematicide application in Bengali lowland rain-fed rice fields infested by M. graminicola resulted in a yield increase of 16 to 20% or about 1 T/ha [14], while yield increases of 12 to 33% and 28 to 87% were observed in M. graminicola-infested upland rice fields respectively in Thailand [15] and Indonesia [16]. However, the chemicals used for RKN control are toxic not only to humans, but also to the environment and have been or are being banned in most countries [17]. Consequently, RKN are an increasing pest problem, prompting the urgent need for alternative and durable management strategies [18].
Vietnam has been one of the world's largest rice exporters since the mid-1990s but is also one of the world's top five rice consumer countries [19]. In contrast with Central and North Vietnam, where farmers grow a large diversity of rice varieties mostly for local needs, the Mekong River Delta area in South Vietnam is under intensive rice production using fewer rice varieties. In this area, three rice crops are produced per year accounting for about 90% of the exported rice [20]. The other intensive rice-growing regions are along the Red River Delta in the North of Vietnam (18% of the national paddy rice production), the North-East and the North-Central Coast. In this regard, it is unfortunate that knowledge concerning M. graminicola is limited to the southern part of the country (Mekong River Delta) and dates back to more than 20 years ago [21].
Only a few studies have been performed to assess the genetic diversity of M. graminicola. Until now, no isozyme polymorphism has been revealed for M. graminicola (e.g., [22]). The Internal Transcribed Spacer (ITS) region rendered a few polymorphisms that were used to determine the phylogenetic relationships among isolates from the USA, India and Bangladesh [23]. Studies of M. graminicola isolated from rice in several countries have revealed diversity among these populations and two pathotypes have been described on the basis of their virulence on different plant species [24]. To date, the ITS is the only genetic marker that has been extensively used on M. graminicola.
In our study, the diversity of M. graminicola was analysed using populations isolated from rice in ten Vietnamese provinces representative of the various rice agro-ecosystems occurring from North to South Vietnam. Their diversity was evaluated for morphometrics of some selected characters, host plant range, reproduction and virulence (measured as root galling), as well as at the genetic level (ITS). In addition, we developed and validated a SCAR marker suitable for the fast and reliable identification of M. graminicola.
2 Material and methods
2.1 Characteristics of the survey sites
The survey was carried out during June and July 2011. Twenty-one root-knot nematode populations were isolated from rice plants in ten Vietnamese provinces representative for the rice agro-ecosystem diversity in Vietnam (Fig. 1). Vietnam has a land area of about 320,000 km2 and a coastline of 3260 km. Three quarters of its territory are covered by hills and mountains with elevations between 100 and 3400 m, while the plain areas include two major river deltas: the Red River Delta in the North and the Mekong River Delta in the South. Vietnam has, in general, a tropical monsoon climate with regional climate variations that are considerable due to the length of the country and its diverse topography. The North has a humid subtropical climate where the annual humidity averages 84%, and the summers are warm and very humid while winters are cold and relatively dry [25]. The North-Central area climate is characterised by a relatively cold winter, a dry and hot west wind, and high rainfall in the second season of the year. In contrast with these areas, the South-Central area has a warm winter, a rainy season that occurs in late summer and an early winter with a particularly low rainfall. Finally, further south, especially in Ho Chi Minh City and in the surrounding Mekong River Delta, the climate predominantly has tropical savannah characteristics with a high humidity, and a distinct wet and dry season [19]. In 2011, annual mean temperature in Vietnam varied from 18 °C to 29 °C. The coldest monthly means ranged from 13 °C to 20 °C in the northern mountainous areas and from 20 °C to 26 °C in the southern areas. As for the hottest periods, monthly means temperatures varied from 25 °C to 30 °C in the northern mountainous part, from 20 °C to 31 °C in the central region, and from 27 °C to 29 °C in the delta Mekong River [26]. Diverse rice agro-ecosystems are present: in the North-East and North-West, as well as in part of the mountainous central region (Da Lat), rice is grown in a rain-fed upland agro-ecosystem whereas a lowland agro-ecosystem is usually found in the deltas of the Red River and Mekong River.

(Colour online.) Map indicating survey sites within Vietnam. Three main climates are observed in this country: a humid subtropical climate in the North with high humidity all year long; a humid subtropical climate with torrential rain from typhoons in the Central part, and a tropical savannah climate in the South. Borders between these regions are indicated in red and green lines, red spots indicate survey sites.
2.2 Sampling
The 21 populations were randomly selected in ten different provinces to cover Vietnamese rice agro-ecosystems diversity. Except in the Mekong River Delta area, where plants were at their ripening stage (grain maturation), all other rice plants sampled were 25- to 50-days old at the vegetative stage (before panicle initiation) or the initiation of the reproductive stage. Plants sampled around Da Lat were usually direct-seeded while most other plants sampled had been transplanted. At each survey site, the rice root systems of 40 randomly collected plants were carefully scanned for the presence of characteristic galls (with terminal hooks) indicating infection by M. graminicola. A representative composite infected root sample was collected, immediately placed in a plastic bag and transported to the laboratory. For each survey site, the number of crop cycles per year, the rice varieties and the plant protection practices were recorded. Samples were labelled and kept at 4 °C in a refrigerator. In the laboratory, galls were dissected using a stereomicroscope and were either transferred to a 0.4 M NaCl solution at 4 °C until nematode extraction was performed or used to confirm the presence of M. graminicola by acid fuchsin staining [27].
2.3 Nematode extraction
Eggs were recovered from infected roots using the hypochlorite extraction method with minor modifications [28]. Infected roots were collected, washed and cut into small (2 cm) segments before being blended for 30 s in 0.8% hypochlorite solution. The mixture was left for 10 min in the hypochlorite solution, before being filtered through 250-μm and 50-μm sieves to remove the plant root tissues. Eggs were recovered on a third 25-μm sieve, rinsed several times with sterile ddH2O and placed on a strainer covered by three damp Kimwipe tissues on a 50-mL beaker filled with sterile ddH2O. After 24 h in the dark at room temperature, the water was removed and replaced with new sterile ddH2O. For the morphometric study, freshly hatched second-stage juveniles (J2) were picked randomly from the bottom of the beaker, placed in a drop of water on a glass slide, killed by gentle heat and covered with a glass cover slip.
2.4 Establishment of subcultures
After nematode extraction, one individual J2-stage nematode of each sample was randomly selected to establish a representative population for each survey site. These subcultures were established in a net house at the Hanoi University of Agriculture on rice cultivar (cv.) IR64 grown in a previously autoclaved sandy soil made up of 50% sand and 50% potting soil and watered every three days in order to conserve a non-saturated soil (50% of soil pore volume filled with water). Distinguishable by their dark gut due to storage of lipids visible under a stereomicroscope, freshly hatched J2s were manually picked and inoculated onto two-weeks-old rice seedlings.
2.5 Morphometric study
J2s of M. graminicola were collected from rice roots from the 21 populations. Population VN3 was lost during the experiment and thus not added in the final analysis. Ten J2s per population were randomly selected for measurement of both body and stylet lengths. The same nematode extraction method (see above) was used to extract J2s from the directly collected field samples and from the populations propagated on the rice cv. IR64 (subcultures) with the aim to compare the variation of body and stylet lengths following nematode reproduction under different conditions (i.e. field vs. net house).
2.6 Nematode reproduction and virulence study
Due to the variability in their ITS sequences among the M. graminicola populations (see below “molecular study” section), eight Vietnamese M. graminicola populations were selected and multiplied on rice cv. IR64 as described above. This subset of populations was used in a net house experiment to evaluate their reproduction and virulence on the rice cv. IR64 grown in a previously autoclaved sandy soil made up of 50% sand and 50% potting soil. Ten replicates of 200 J2s of each population (Pi: initial population density) were inoculated on one-week-old rice seedlings growing under a non-saturated water regime. Two days after inoculation, pots were flooded intermittently three times per week to imitate the water regime of the summer-irrigated lowland rice-growing cultivation. The root systems were collected and the eggs extracted using the hypochlorite extraction method after two nematode life cycles. As confirmed by acid fuchsin staining, the first eggs appeared in infected roots 18–20 days after infection and continued to be produced by each female during ten days (data not shown). Therefore, to ensure a significant measurement of the reproductive factor for this experiment, one life cycle was considered as 30 days after infection and 60 days for two life cycles. After two weeks in the dark at room temperature, the hatched J2s were collected to assess the final population density (Pf). The reproductive factor (Rf) was calculated as the Pf/Pi ratio where plants with Pf/Pi ≤ 1 are considered as resistant and Pf/Pi > 1 as susceptible according to the pioneering work on rice-nematode resistance performed by Soriano et al. [29].
2.7 Nematode host plant range study
The host plant range of the 21 Vietnamese populations propagated on rice cv. IR64 was investigated. Two experiments each with 10 replicates were carried out with the following potential host plants: tomato (Solanum lycopersicum L.) cvs Money Maker and Rutgers, tobacco (Nicotiana tabacum L.), maize (Zea mays L.) cv. MX2, rice (Oryza sativa L.) cv. IR64, Welsh onion (Allium fistulosum L.), common onion (A. cepa L.), garlic (A. sativum L.), peanut (Arachis hypogaea L.) cv. V5, green bean (Vigna radiata L.), mung bean (V. cylindrica L.) and soybean (Glycine max L.) cv. DT84. Two hundred freshly hatched J2s were inoculated on plants in full vegetative growth stage with a well-developed root system (three-weeks-old for rice plants). Plants were grown in 10 × 10 cm pots filled with previously autoclaved sandy soil made up of 50% sand and 50% potting soil and watered every three days in order to conserve a non-saturated soil. Plants were randomly distributed in the experiments. Nematode-infected plants were maintained in a net house for 60 days before the roots were harvested and examined for the presence of root galls. Rating of root galling was assessed as follow:
- • – = no galls;
- • + = 1 gall;
- • ++ = 2–10 galls;
- • +++ ≥ 10 galls.
The presence or absence of nematodes (adult, juvenile and egg stages) was confirmed by observation under the stereomicroscope of acid fuchsin stained roots [27].
2.8 Statistical analyses
Statistical analyses were done using the statistical software R [30]. The stylet lengths were transformed by 1/(stylet length) to obtain a normal distribution. All other requirements were answered. ANOVAs were performed on the body length, the stylet length and the reproductive factor (Rf). Differences among populations (or locations) for body length, stylet length and Rf were analysed by multiple comparisons using the test of Tukey's Honest Significant Differences (Tukey HSD). A non-parametric Spearman's rank correlation test was performed between the body lengths and stylet lengths. The significance level was considered at 0.05 indicating that the observed result would be highly unlikely under the null hypothesis.
2.9 DNA analyses
2.9.1 Development of a SCAR marker specific to M. graminicola
To develop a Sequence Characterised Amplified Region marker (SCAR), two populations of M. graminicola originating from Brazil [22] and the Philippines [29] as well as the 21 Vietnamese populations were used (for details on the SCAR marker development: [see Supporting information], [53]). Specific primers were designed (SCAR-MgFW: 5′-GGGGAAGACATTTAATTGATGATCAAC-3′ and SCAR-MgRev: 5′-GGTACCGAAACTTAGGGAAAG-3′). Amplifications were performed using the following conditions: 1 min at 95 °C, 30 cycles of 30 s at 95 °C, 30 s at 60 °C and 1 min at 68 °C followed by a final extension step of 10 min at 68 °C. For each nematode population, genomic DNA was purified from aliquots of 100-μL eggs by a phenol–chloroform method [31]. Total genomic DNAs (gDNA) from M. incognita, M. javanica and M. arenaria were kindly provided by P. Castagnone (INRA Antibes, France) and gDNA from M. exigua and M. paranensis were kindly provided by F. Antony and L. Vilain (IRD Montpellier, France). DNA extraction and PCR from single J2 was performed according to the protocol described for “ITS amplification and sequencing” with the “Phusion High-Fidelity DNA Polymerase” (Thermo Fisher Scientific Inc.) and the following conditions: 98 °C for 30 s; 35× (98 °C, 10 s; 60 °C, 30 s; 72 °C, 40 s); 72 °C for 10 min.
2.9.2 ITS amplification and sequencing
Individual J2 nematodes were manually collected using a stereomicroscope and transferred to a 0.2-mL polymerase chain reaction (PCR) tube containing 10 μL of sterile ddH2O. An equal volume of 2 × lysis buffer containing 20 mM Tris–HCl pH 8.4, 100 mM KCl, 3 mM MgCl2, 0.9% Tween 20 and 2 mM DTT was added. Proteinase K to a final concentration of 20 μg/mL was added and the tube was placed in a water-bath at 50 °C for 2 h. To partially inactivate the proteinase K, reactions were incubated for 15 min at 100 °C. Lysate was used immediately or stored at –20 °C. Primers rDNA2 (5′-TTGATTACGTCCCTGCCCTTT-3′) and rDNA1.58s (5′-ACGAGCCCGAGTGATCCACCG-3′) were used for ITS-1 amplification of rDNA genes [23,32,33]. The ITS was amplified by PCR in a final volume of 50 μL containing 5 μL of crude DNA extract, 500 nM of each PCR primer, 200 μM each dNTP, 50 μg of BSA, 10 μL 5 × Phusion buffer and one unit of the proof-reading enzyme “Phusion High-Fidelity DNA Polymerase” (Thermo Fisher Scientific Inc.). The following PCR program was used: 98 °C for 30 s; 35 × (98 °C, 10 s; 54 °C, 30 s; 72 °C, 40 s); 72 °C for 10 min. PCR products were gel-purified and eluted in 30 μL ddH2O using a gel purification kit (Qiagen). Seven microliters of the product were used to add an A-tail by mixing it with 5 units of Taq DNA Polymerase, 1 μL 10 × Taq buffer (Biolabs, France), 0.2 mM dATP and incubating for 30 min at 68 °C. ITS fragments were cloned into the pGEM®-T Easy Vector (Promega) and sent for sequencing using standard procedures and M13 primers (Cogenics, UK).
2.9.3 Sequence alignment and phylogenetic analyses
Following protocols described in previous M. graminicola phylogenetic analyses [23], a 386-bp region including a portion of the 18S rDNA gene (151 bp), the complete ITS-1 and part of the 5.8S rDNA gene, were chosen. For 12 nematode populations (at least one population/prospected province), the 386-bp DNA region was amplified from ten independent single J2 nematodes. For Lao Cai province, two populations (VN17 and VN18) were selected since this mountainous province includes different agro-ecosystems. For each population, the ten sequences were compared and the consensus sequence was deposited in the GenBank database. We also sequenced and deposited in the GenBank database this same DNA region from two M. graminicola accessions collected in Brazil (MgB) and the Philippines (MgP) [22,29].
We imported from GenBank ITS sequences of other M. graminicola accessions. On the June 30th, 2014, 45 sequences were downloaded and were aligned with our sequences using ClustalO [34]. The quality of sequences from GenBank was checked using the ribosomal 18S gene that is expected to be highly conserved at the intraspecific level in Meloidogyne (e.g., [35]) and sequences that show at least three mismatches in this gene were discarded because likely correspond to pseudogenes, sequences with PCR errors or incorrectly edited sequences.
The Phylogeny.fr web platform [36] was used to build a phylogenetic tree from nematode ITS sequences download from GenBank or generated in this study (accession numbers on Fig. 5). This platform offers a predefined pipeline using MUSCLE, Gblocks, PhyML and TreeDyn that outputs the corresponding phylogenetic tree. Phylogenetic trees were generated by the maximum-likelihood method using the default parameters. Each phylogram was bootstrapped 500 times to assess the degree of support for the phylogenetic branching, indicated by the optimal tree. A haplotype network was also reconstructed based on our ITS sequences. A reduced-median network was built using Network v.4.112 [37]. Indels were excluded from the analysis and our analysis was finally based on 20 nucleotide substitutions (among which five are parsimony informative sites).
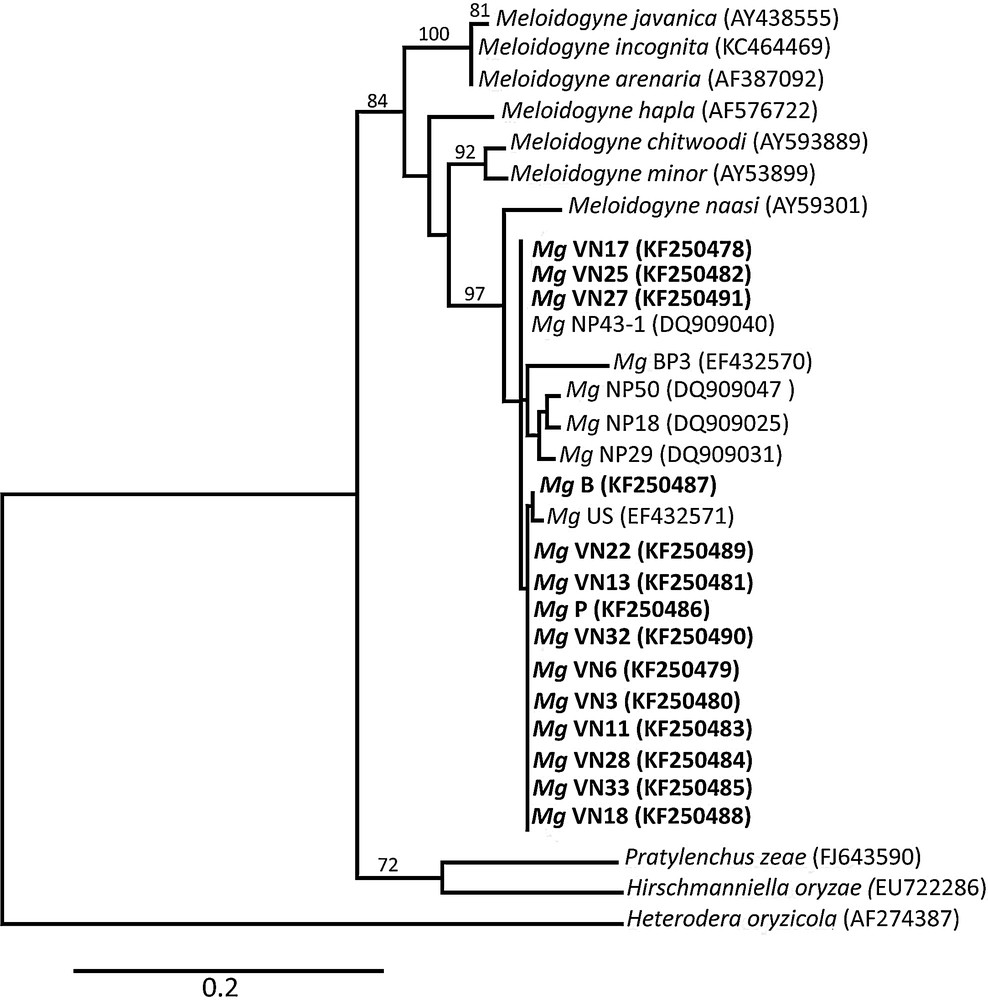
Evolutionary relationships among plant–parasitic nematode spp. ITS lineages are estimated using maximum-likelihood. Branches with bootstrap support > 65% are indicated. The scale bar denotes 0.2 substitutions per nucleotide position. Mg: Meloidogyne graminicola; VN: Vietnamese populations; B: Brazilian population and P: Philippines population (this study). NP: Nepalese populations; BP: Bangladesh population and US: USA Florida population. Newly obtained sequences are indicated in bold letters.
3 Results
3.1 Characteristics and sampling of survey sites
All survey sites examined were infested with M. graminicola (Fig. 1). The frequency of occurrence of M. graminicola was higher on the bounds of the surveyed fields, near the paths, than in the middle of the fields (data not shown). The nematodes were most abundant in younger plants AQ (one month after transplantation) than in older ones. In addition, M. graminicola was often found in roots of several weed species (e.g., Cyperus iria L.) at the edges of the rice fields as previously described [38]. During the first month after transplantation, infested fields showed symptomatic patches with lower plant density and a delay in rice development (Fig. 2). In these patches, the rice root systems showed the typical hook-like root galls caused by M. graminicola. These typical galls were detected on all rice developmental stages and in all rice agro-ecosystems surveyed.
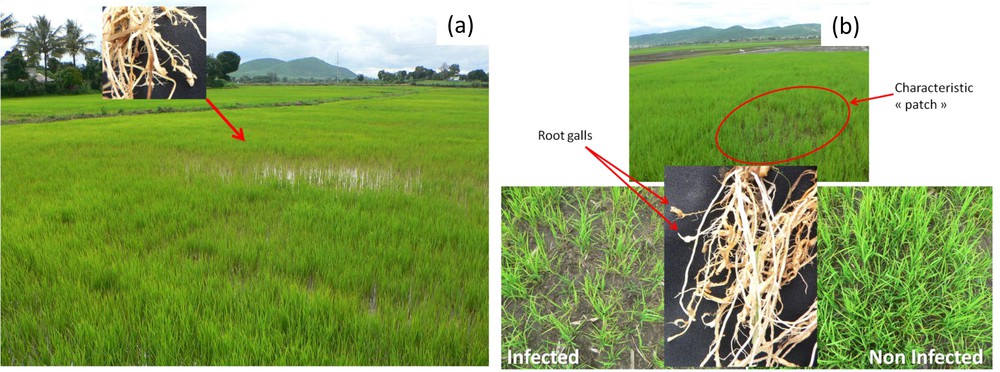
(Colour online.) Symptoms in Meloidogyne graminicola-infested fields. a: lowland rice; b: upland rice. Rice root systems showed typical root gall formation with a hook-like tip (root galls).
3.2 Nematode morphometric study
Our results indicate that both body length and stylet length are highly similar within each collected population as reflected by the small standard error when measuring ten J2 nematodes per population (Table 1). Populations showed significant (P < 0.05) variability in terms of body and stylet lengths, however, the most significant difference was observed for the body length (Table 1). J2 body length ranged from 367 to 501 μm (average 440 μm), the population VN18 having the longest body length (on average 481 μm) and population VN13 the smallest (on average 397 μm). J2 stylet length ranged from 11.7 to 17.3 μm (average 14.3 μm), the population VN12 having the longest (on average 15.6 μm) and population VN33 the smallest stylet length (on average 12.8 μm). No correlation was found between the body length and the stylet length (data not shown). For example, population VN18, which has the longest body length, has an average stylet length. Populations collected in mountainous areas were significantly (P < 0.01) longer than those collected from rice growing in valleys or deltas (respectively, on average 452 μm vs. 435 μm and 439 μm; Table 1).
Origin and morphometric measurements of Vietnamese second-stage juveniles (J2) M. graminicola collected in rice fields. All measurements are in μm. Values are the mean ± SE. Columns indexed by the same letter are not significantly different according to Tukey's honestly significant difference test (P < 0.05).
| Population code | Geographical region | Agro-ecological region | Province | Body length (μm) | Stylet length (μm) | ||||||
| Range | Mean | SE | Range | Mean | SE | ||||||
| VN6 | Delta | Mekong River Delta | Dong Thap | 400–501 | 443 | 12.3 | bcd | 12.44–19.03 | 15.02 | 1.3 | ab |
| VN11 | Delta | North-Central Coast | Nghe An | 375–470 | 436 | 8.6 | bcd | 12.93–17.07 | 14.95 | 0.4 | ab |
| VN12 | Delta | North-Central Coast | Nghe An | 390–495 | 429 | 10.2 | cde | 13.41–17.31 | 15.58 | 0.4 | a |
| VN13 | Delta | Red River Delta | Ha Noi | 385–415 | 397 | 3.1 | e | 12.93–17.32 | 14.83 | 0.5 | ab |
| VN14 | Mountain | North-east | Lao Cai | 367–460 | 415 | 8.2 | de | 12.68–15.85 | 14.15 | 0.3 | ab |
| VN15 | Mountain | North-east | Lao Cai | 430–490 | 452 | 6.2 | abcd | 14.15–17.32 | 15.30 | 0.3 | a |
| VN17 | Mountain | North-east | Lao Cai | 440–490 | 464 | 5.2 | ab | 12.68–15.85 | 14.41 | 0.3 | ab |
| VN18 | Mountain | North-east | Lao Cai | 450–500 | 481 | 4.3 | a | 13.41–16.10 | 14.70 | 0.2 | ab |
| VN22 | Valley | North-east | Bac Can | 402–478 | 441 | 7.6 | bcd | 12.44–14.88 | 14.04 | 0.2 | ab |
| VN24 | Valley | North-east | Bac Can | 395–465 | 429 | 7.3 | cde | 12.44–16.34 | 13.94 | 0.4 | ab |
| VN25 | Mountain | North-west | Son La | 400–470 | 447 | 7.8 | abcd | 12.80–15.85 | 14.30 | 0.3 | ab |
| VN26 | Delta | South-Central Coast | Quang Nam | 405–462 | 432 | 7.4 | bcd | 12.20–14.88 | 13.78 | 0.3 | ab |
| VN27 | Delta | South-Central Coast | Quang Nam | 436–482 | 458 | 4.9 | abc | 12.44–16.34 | 14.20 | 0.4 | ab |
| VN28 | Valley | North-Central Coast | Hue | 411–490 | 447 | 7.6 | abcd | 12.68–16.59 | 14.41 | 0.4 | ab |
| VN29 | Valley | North-Central Coast | Hue | 425–458 | 437 | 3.3 | bcd | 12.68–17.07 | 14.24 | 0.4 | ab |
| VN30 | Valley | North-Central Coast | Hue | 419–445 | 434 | 2.5 | bcd | 12.20–17.07 | 14.24 | 0.4 | ab |
| VN31 | Valley | North-Central Coast | Hue | 421–465 | 445 | 5.0 | bcd | 12.68–14.63 | 14.04 | 0.2 | ab |
| VN32 | Delta | Mekong River Delta | Ho Chi Minh | 395–472 | 433 | 9.6 | bcd | 12.20–14.15 | 13.41 | 0.2 | ab |
| VN33 | Delta | Mekong River Delta | Ho Chi Minh | 394–462 | 429 | 6.6 | bcde | 11.71–14.63 | 12.76 | 0.3 | b |
| VN34 | Delta | Mekong River Delta | Ho Chi Minh | 434–476 | 456 | 4.4 | abc | 12.20–14.63 | 13.54 | 0.2 | ab |
A set of populations showing the most extreme body or stylet lengths (VN13, VN18, VN33) as well as another set with intermediate body and stylet lengths (VN6, VN11, VN17, VN22, VN27, VN30) were propagated again during six months and re-extracted from rice cv. IR64 growing in the net house. Significant variability was found for the body and the stylet lengths and the ranking previously observed among the populations was conserved: VN13 remained the population having the smallest body length (on average 406 μm), VN18 having a relatively long body length (on average 483 μm) and VN33 having the smallest stylet length (on average 13 μm; Table 2). For most populations, the body length was slightly longer when the nematodes were propagated on rice cv. IR64 in the net house than when collected directly from the field, except for VN17 of which the body length was significantly (P < 0.05) shorter in the net house population compared with the field population (–9%; Table 2). Population VN27 showed the highest significant increase in body length when propagated in the net house (+ 4.5%).
Morphometric measurement of Vietnamese second-stage juveniles (J2) M. graminicola propagated on rice cv. IR64 in a net house. Values are the mean ± SE. Columns indexed by the same letter are not significantly different according to Tukey's honestly significant difference test (P < 0.05).
| Population code | Body length (μm) | Stylet length (μm) | ||||||||
| Range | Mean | SE | Variation (in %)a | Range | Mean | SE | Variation (in %)a | |||
| VN6 | 405–471 | 447 | 7.3 | abc | 1.0 | 12.80–15.85 | 14.22 | 0.3 | ab | –5.8 |
| VN11 | 411–490 | 446 | 8.1 | bc | 2.3 | 12.88–16.59 | 14.49 | 0.4 | a | –3.1 |
| VN13 | 350–490 | 406 | 13.6 | d | 2.4 | 12.68–14.88 | 14.19 | 0.2 | ab | –4.4 |
| VN17 | 405–445 | 425 | 4.4 | cd | –9.1 | 12.32–14.72 | 13.52 | 0.3 | ab | –6.7 |
| VN18 | 450–510 | 483 | 5.5 | a | 0.4 | 13.48–16.10 | 14.74 | 0.2 | a | –0.0 |
| VN22 | 386–468 | 435 | 7.4 | cd | –1.5 | 13.44–16.69 | 14.08 | 0.1 | ab | 0.4 |
| VN27 | 436–512 | 479 | 6.9 | ab | 4.3 | 12.44–15.47 | 13.70 | 0.3 | ab | –3.6 |
| VN30 | 405–485 | 440 | 8.4 | cd | 1.5 | 12.20–14.88 | 13.83 | 0.3 | ab | –3.2 |
| VN33 | 394–480 | 435 | 7.6 | cd | 1.3 | 11.71–14.63 | 13.00 | 0.3 | b | 1.8 |
a Variation corresponds to the difference in % between field collected population and population propagated under controlled conditions in the net house. All measurements are in μm.
3.3 Nematode reproduction study
Under our experimental net house conditions, an acid fuschin-staining assay revealed that all M. graminicola populations took 18 to 20 days to complete their life cycle on cv. IR64 (data not shown). The reproductive factor (Rf = Pf/Pi) was calculated for populations collected in various regions including at least one from each ITS haplotype (see below). After two life cycles, all populations were highly reproductive on rice cv. IR64 with a Rf value ranging from 11 to 19 depending on the population (Fig. 3). The VN17 population collected from the mountains had the lowest reproduction factor on IR64 (P < 0.05). However, there was no significant correlation between the Rf and either geographic origin of the populations, soil type or crop rotation system (data not shown).
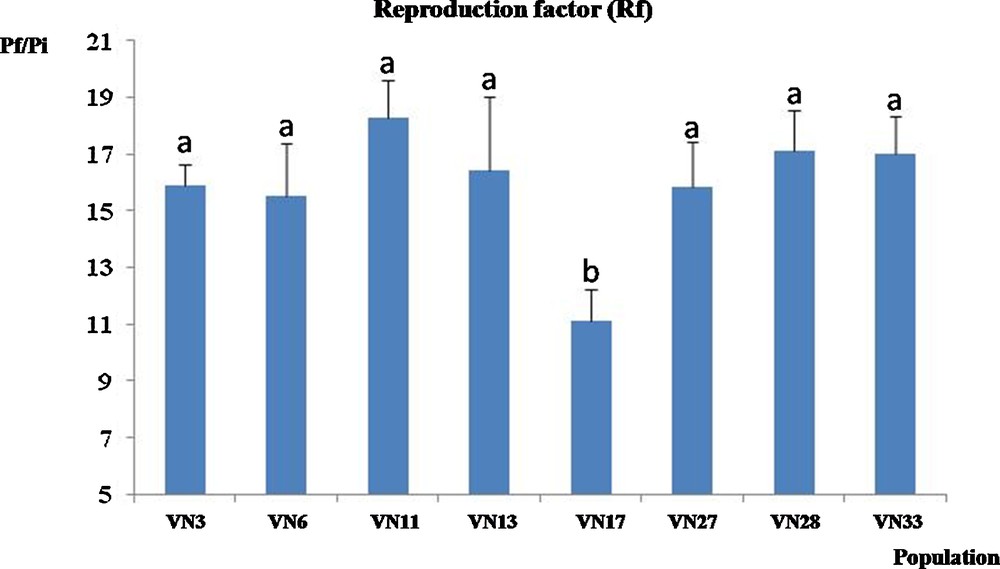
(Colour online.) Reproduction factor (Rf) of Vietnamese Meloidogyne graminicola populations on rice cv. IR64. Rf corresponds to the Pf/Pi ratio. Values are the mean ± SE. Columns indexed by the same letter are not significantly different according to Tukey's honestly significant difference test (P < 0.05).
3.4 Nematode host plant range study
All 21 Vietnamese M. graminicola populations induced the characteristic galls at the rice root tips with a hook-like shape on rice cv. IR64 under net house growing conditions. When the infection is severe, the galls can become globular. None of the populations were able to multiply on the tomatoes cvs Rutgers or Money Maker or on the tobacco, peanut, maize, green bean, mung bean or soybean cultivars included in the experiments [see Supporting information]. However, all M. graminicola populations were able to multiply on rice cv. IR64 and cultivars of garlic and onion [see Supporting information] with on average 50 to 100 galls per plant (data not shown).
3.5 Development of a SCAR marker specific to M. graminicola
A specific SCAR maker to M. graminicola was developed [see Supporting information] and the sequence of the amplification product was deposited in the GenBank database (under accession number KF499563). Using “nucleotide collection” from NCBI, BLASTN search did not reveal any significant similarity with other sequences. However, a BLASTn search against the M. incognita genome (http://meloidogyne.toulouse.inra.fr/blast/blast.html) revealed a homologous region on contig MiV1ctg921 within the predicted Minc15141 gene. Clustal Omega alignment (http://www.clustal.org/omega/) showed that 63.5% of the nucleotides were identical between MiV1ctg921 fragment and the M. graminicola SCAR amplicon [see Supporting information]. These SCAR primers did not allow amplification either from M. incognita, M. arenaria, M. javanica, M. exigua or from M. paranensis genomic DNA (Fig. 4a). A specific 640-bp amplification was obtained with the two M. graminicola populations from the Philippines and Brazil (Fig. 4a), and with the 21 Vietnamese populations (Fig. 4a and c). The consensus sequence between M. graminicola populations is almost identical with only two SNP at position 329 (C or T) and 510 (A or G) inside the 640-bp fragment (data not shown). Using gDNA extracts from a single J2 nematode and a single male, the SCAR primers allowed amplification of the 640-bp specific band (Fig. 4b), thus demonstrating the sensitivity of the detection.
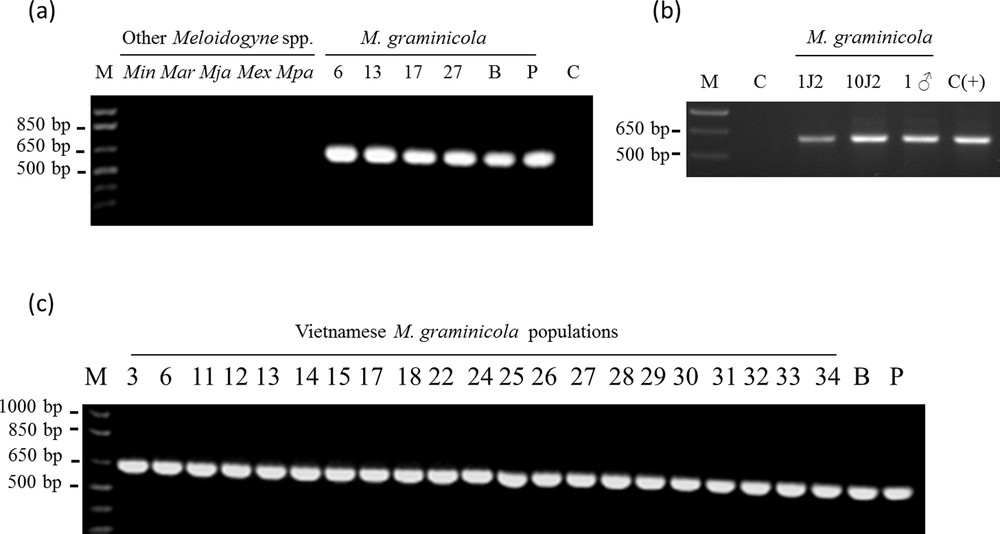
Amplification patterns for 28 Meloidogyne spp. populations generated with the specific SCAR-MgFW/Rev primers. a: Specific SCAR-PCR amplicon generated by using different Meloidogyne spp. genomic DNA; b: M. graminicola SCAR-PCR amplicon from one second-stage juvenile (J2), ten J2s and one male using the SCAR primers; c: specific SCAR-PCR amplicon generated by using genomic DNA of the 21 Vietnamese populations and two different M. graminicola populations as control. M, 1Kb plus Ladder (Invitrogen); C, control reaction without template DNA; C(+), control reaction using 10 ng of M. graminicola gDNA. Min: M. incognita; Mar: M. arenaria; Mja: M. javanica; Mex: M. exigua; Mpa: M. paranensis; B: M. graminicola (population from Brazil); P: M. graminicola (population from the Philippines). 3–34: 21 Vietnamese M. graminicola populations (VN).
3.6 ITS amplification and phylogenetic analysis
For each analysed population, the sequences obtained from all ten J2 nematodes were identical. Based on a BLAST search, all populations matched with M. graminicola population BP3 (GenBank accession EF432570) [23] with a score ranging from 94% identity, (E value 3e−168), to 95% (E value 5e−176). The Vietnamese M. graminicola populations were clearly different compared with the other Meloidogyne species that are spread in Vietnam and could infect rice (Fig. 5), including M. javanica, M. arenaria and M. incognita (i.e. VN3 and M. incognita have sequence identities of only 86%). The ITS sequences of the analysed Vietnamese Meloidogyne spp. thus all belong to M. graminicola.
3.7 Haplotype network analysis
A haplotype network was constructed using the ITS sequences generated for this study and those imported from the NCBI database (Fig. 6). Twenty-three sequences from GenBank with more than two nucleotide mismatches in the 3′-extremity segment (151 bp) of the 18S gene were first excluded. This region was invariable in our sequences. The 18S gene is expected to be highly conserved at the intraspecific level (e.g., [35]) and the presence of numerous polymorphisms in this region suggests that most M. graminicola ITS sequences retrieved from GenBank were either incorrect (due to PCR errors or incomplete sequence editing) or correspond to pseudogenes. For this reason, these sequences were not considered further in the network analysis. Most of the available M. graminicola ITS sequences have been isolated from Nepalese populations (31 accessions [see Supporting information], [23]), but 19 of them were discarded based on our criteria. Consequently, available sequences from Nepal have to be considered with caution, and branches of unique sequences were represented in the network with dotted lines (Fig. 6).

(Colour online.) Reduced-median network of the ITS reconstructed with Network v. 4.112 [37]. The network is based on 20 substitutions (of which five are parsimony informative). Each haplotype is numbered and represented by a pie chart. The size of the pie chart is proportional to the number of accessions with a particular haplotype. The origin of haplotypes is indicated by specific colour. Eight haplotypes from Nepal (i.e. 6, 7, 9–13, 15) were observed once and suspected to be misleading sequences (see results). Their relationships to other haplotypes were thus represented with dotted lines.
Fifteen different haplotypes were identified, each represented by one to fourteen M. graminicola populations (Fig. 6) [see Supporting information]. Four ITS types were detected in Vietnam (Fig. 6). Only one of them has never been observed in previous studies. Each Vietnamese ITS types are distinguishable from the others by at least two nucleotide substitutions. However the phylogenetic relationships between these four haplotype are not resolved suggesting either homoplasy (recurrent mutations on the same sites) or recombination between distinct types. Haplotype no 1 is the most frequent and is connected to eight other haplotypes including the Brazilian accession. These features suggest that this haplotype is the ancestral state in South Asia. No clear pattern of geographic distribution of haplotypes was observed. Indeed, three Vietnamese haplotypes were shared with accessions from Nepal, India, the Philippines or China.
4 Discussion
4.1 On the ascendency of M. graminicola among RKN species in Vietnamese rice fields
Although several Meloidogyne spp. have been reported on rice plants [39,40], our survey only reveals M. graminicola in Vietnamese rice agro-ecosystems. Since M. incognita and M. javanica, have been found in Vietnam on other crops, such as bananas [41], it was surprising to find only M. graminicola in lowland and upland rice. It has been shown previously that M. graminicola is highly adapted to the rice-growing conditions found in flooded soils [42]. However, competition with other Meloidogyne spp. must occur in upland rice. The exclusive presence of M. graminicola in these agro-ecosystems is therefore particularly intriguing. Similar observations were made in Nepal, Bangladesh, South Vietnam, Indonesia and the Philippines where only M. graminicola was reported in association with rice root systems [7,14,16,21,23]. Further studies are required to determine whether Vietnamese M. graminicola populations could outcompete other Meloidogyne spp. in the rice root system or whether its predominant occurrence is the consequence of particular agronomic practices.
The presence of M. graminicola in roots of several weed species could also be important for the management of this nematode species. It was previously reported that M. graminicola has a wide host plant range that includes many of the common weeds of rice fields [38,43]. It is important to determine whether weed species associated with rice fields could play a role as significant reservoirs of this rice pathogen (particularly on the edge of fields or near the paths).
4.2 Assessment of the morphological variability among Vietnamese M. graminicola populations
Quantitative characters, such as the tail shape and tail length [44] can be used for the identification of several Meloidogyne species [45] but the body and stylet lengths of M. graminicola are very variable and rule out the use of these traits [24]. However, comparison of body and stylet length among different M. graminicola populations (Nepal, India, Bangladesh, Thailand and the USA) has already shown intraspecific variations [24,45]. Comparison of all Vietnamese populations examined showed significant variations in body and stylet length. These variations were observed on freshly hatched J2 nematodes isolated from different rice cultivars growing in different agro-ecosystems. No correlation was found between the body and the stylet length, which is in agreement with previous observations made on diverse M. graminicola populations [24,45]. In spite of the fact that all populations were propagated at the same time for several cycles on rice cv. IR64 under net house conditions, significant differences in the body and stylet length of all populations examined were conserved when compared to populations directly isolated from the field. However, population VN17, which was isolated from the mountains of North-West Vietnam showed a significant diminution of its body length when cultivated on cv. IR64 under net house conditions (–9%) and another population, VN27, isolated near the ocean was significantly longer after propagation on cv. IR64 (+ 4.5%). Taken together, these observations suggest that the differences observed among populations are mainly determined at a genetic level, but environmental factors (i.e. soil type, climate, plant host, etc.) may also have an effect on some morphometrical characteristics of J2 nematodes. Interestingly, populations from the mountains have a smaller body shape compared to the populations from the delta rivers. We cannot determine if this heritable difference is a consequence of soil characteristics, climate, rice genotype or abiotic or biotic stress(es) but a selective pressure seems to occur leading to populations of different body shape.
4.3 Assessment of the pathogenicity variability among Vietnamese M. graminicola populations
Different M. graminicola populations showing variability in host plant specificity and reproduction have been previously isolated [23] and two pathotypes have been identified with one pathotype maintained on O. sativa that can infect and reproduce in rice cv. BR11 whereas the other pathotype which is maintained on the weed C. rotundus is not able to reproduce on this rice cultivar but as the other pathotype it can reproduce on wheat [24]. The concept of pathotype (also named race) was defined by Sasser [46] as a set of populations sharing distinctive physiological characters that result in their ability to reproduce, or not, on selected differential hosts. In our study, the host range used did not reveal significant differences among the M. graminicola populations included in the experiments. Populations were able to propagate on rice and onion, which is in agreement with a previous study [47]. However, the population VN17, isolated from the mountains in North Vietnam from a local rice variety, showed a normal reproduction rate on onion but a significantly lower reproduction rate on the rice cv. IR64 under net house conditions. One M. graminicola pathotype identified by Pokharel et al. [24] was not able to propagate on different rice cultivars such as the two US O. sativa cultivars, Labelle and LA110, as well as the BR11 popular rice variety from Bangladesh [24]. We cannot exclude that differential hosts could be found for some populations if using a larger number of hosts, especially to investigate the reproduction of VN17 in more depth.
All Vietnamese M. graminicola populations were not able to reproduce on tomato, green bean or soybean. In contrast these three species were reported as hosts for M. graminicola [43] but no information has been given concerning the origin of the M. graminicola population tested in this host range study. In addition, all Vietnamese populations were not able to reproduce on maize. This is in contradiction with observations done on other M. graminicola populations (from India, Bangladesh and the USA) that all could reproduce on maize [24] but in agreement with the results of a host range study performed by Yik and Birchfield [38] using three different maize cultivars and a M. graminicola population from the USA (Louisiana). Several hypotheses can be proposed to explain the difference in the host range observed between the Vietnamese and these other M. graminicola populations. The maize cultivar that was used in our experiment is different to the one used by Pokharel et al. [24]. So, we cannot exclude the fact that the two different cultivars used in each study present different levels of susceptibility to the nematode infection. A second hypothesis is that the water regimes and soil types were different to the ones used in other studies. Indeed, success in M. graminicola infection is known to be highly correlated with the water regime [8,10]. Unfortunately, in many studies detailed information on the water regime is lacking. A third possibility is that the Vietnamese M. graminicola populations constitute a new pathotype, distinct from those tested by Pokharel et al. [24]. Unfortunately, we did not have any of these other M. graminicola populations to compare them in a host range test to support our hypothesis that the Vietnamese populations could constitute a new pathotype of M. graminicola. Further experiments will have to be performed to answer this question. However, extreme care should be taken not to move new or more damaging M. graminicola pathotypes from the one country or region to another country or region.
4.4 Diagnostic molecular markers to identify M. graminicola and future prospects for intraspecific identification
Using SCAR and ITS DNA markers, we were able to clearly identify M. graminicola. We developed a SCAR marker that allows fast identification of M. graminicola from a single J2 and could be used as a diagnostic tool. Sequencing of SCAR-PCR product from different populations revealed that this sequence is weakly polymorphic inside the M. graminicola species (data not shown) and so constitutes a stable substantial DNA marker. This marker will support RKN management, since agricultural practices are key determinants to manage the build-up of nematode field populations and should be adapted to each Meloidogyne species [42].
In addition, ITS sequence can be used to reveal intraspecific variability among M. graminicola populations [23,24]. Sequence polymorphisms at the level of the ITS sequences allowed us to define four haplotypes from Vietnamese populations. The nuclear rDNA genes and ITS sequences have been found as good nuclear markers that could be used in the identification of several Meloidogyne spp. [23,33]. However, the polymorphism of these markers is limited in comparison with mitochondrial DNA markers [4,48]. Recent studies suggest that nucleotide variations could be found in sequences from the M. graminicola mitochondrial amplification product ([49]; Bellafiore et al., unpublished data). We postulate that more intraspecific molecular diversity exists but new M. graminicola specific mitochondrial DNA markers have to be developed to reveal it. Access to more DNA markers and M. graminicola populations from the rest of the world would certainly improve the strength of the analysis and address the question of the identification of M. graminicola diversity. Mitotic parthenogenetic RKN have an apparent genetic homozygosity whereas amphimictic RKN present more heterogeneity [50]. As M. graminicola belongs to RKN species with a meiotic facultative parthenogenetic reproduction mode [2], which is an intermediate between the amphimictic and mitotic parthenogenetic modes, this nematode species could present more heterogeneity than the mitotic parthenogenetic RKN. If ITS could be used to identify M. graminicola, further markers including microsatellites need to be developed to explore the genomic intraspecific M. graminicola diversity. In particular, new mitochondrial and nuclear markers could help to investigate the evolutionary history of this species (phylogeography) but also to study the dynamic of their populations (e.g., gene flow, population variation size, or importance of each breeding system in variable environmental conditions).
5 Concluding remarks
We have studied the diversity of M. graminicola Vietnamese populations using the data obtained from some selected morphometric traits, host range, reproduction and virulence experiments and developed a DNA marker. Altogether, our observations suggest that Vietnam could be a hotspot for the diversity of M graminicola. As suggested in a previous study conducted in South Vietnam [21], we demonstrated here that M. graminicola is spread in fact all around Vietnam. Characteristic M. graminicola-induced terminal root galls with hook-like shape were found in all rice fields surveyed. A combination of high climate and environmental diversity, high intensity of rice production with three crop cycles per year in several areas, and the high diversity of rice cultivars used by Vietnamese farmers could create favourable conditions for the presence of a large M. graminicola genetic diversity. Further DNA markers need to be developed to continue to explore this apparent diversity and identify the ancestral origin of M. graminicola. Vietnam has already been identified as one of the origins of diversity of several important plant pathogens including viruses from the genus of Begomoviruses and Potyviruses found on a wide range of crops and weeds [51,52] as well as the rice blast fungus Magnaporthe oryzae [20].
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
The authors thank all local farmers of the surveyed regions for their permission to take samples from their fields and for answering our questions during the survey. We thank J. Aribi, C. Viet Ha, H. Nguyen Thi Mai and D. Nguyen Tai for technical assistance and helpful discussions. The authors are grateful to Luanne Maurice for reviewing the manuscript before submission.
Sources of funding: Our work was funding by the Global Rice Science Partnership (GRiSP) Grant (Menergep Project) and the BIOASIA project from the French Ministry of Foreign Affairs and International Development (#4764). GB is supported by the LABEX entitled TULIP managed by the “Agence nationale de la recherche” (ANR-10-LABX-0041).
Appendix A Accession numbers
All ITS sequences obtained in this study from Meloidogyne graminicola have been deposited in the GenBank database under the accession numbers KF250478–KF250491. The 655-bp SCAR N10 fragment amplified from the Philippines population has been deposited in the GenBank database under accession number KF499563.


