1 The cophylogeny: A fundamental approach for investigating evolutionary trends within host-parasite associations
Evolutionary biology is a multidisciplinary approach that seeks to unveil the patterns and processes that shape the phenotypic and genetic diversity. Although organisms may be studied at different levels of their biology, they can also be investigated at different scales of time, with macro evolution involving the largest scales and micro evolution the smallest. Species have evolved through a combination of different processes to fit their changing environments. Many authors are interested in exploring all the features that influence evolution of species, but also how they interact with closely associated non-related species. In specific cases, such as in host-parasite systems, the environment for both interacting species is part of the other living species. Coevolution is a term that was first defined by Ehrlich and Raven [1] to describe changes in one species that may result from evolutionary changes in another one and vice-versa, thus reflecting coadaptation within closely interacting species. For Brooks and McLennan [2], the primary definition of coevolution was too restrictive because it only referred to cases of reciprocal adaptive changes between ecologically interacting species. They gave a broader definition that encompassed coadaptation, for the degree of mutual modification, and cospeciation for the degree of mutual phylogenetic association. Within a systematic framework, phylogenies are very useful for investigating evolution of interacting species, particularly within host-parasite associations. Because cospeciation has been further used in a narrower sense to design synchronous host-parasite divergences [3], this approach is now referred as cophylogeny [4] and has an explicit terminology to account for concordances and conflicts between systematic of hosts and their parasites [5]. Examination of the different levels of fidelity between host and parasite branching patterns thus provides a powerful tool to inspect patterns and processes of parasite diversification over host evolution and geological times.
With the advance of molecular techniques [6–8] and cophylogenetic tools (reviewed in [9]), it is now possible to compare host and parasite distance trees and to explore all kind of evolutionary events that account for parasite evolution. Four main categories of events have been recognised [5] (Fig. 1). Firstly, cospeciation illustrates analogous cladogenetic events within hosts and their associated parasites. Divergences within an ancestral host and its parasite are synchronous over geological times even if host speciation does not necessary entail parasite speciation. However, hosts and their parasites may experience codivergence due to the same causal process, as it is exemplified in biogeographic vicariance where two populations of the same species may diverge after being isolated following the emergence of a geographical barrier. Secondly, duplication refers to parasite speciation within ancestral host without host speciation. Consequently, two or several parasite species forming one monophyletic group may be found within the same host species. This can be observed when parasites colonize microhabitats that, ecologically, are closely related, such as intestinal or branchial cavities. Thirdly, horizontal transfer or host switching may occur if a parasite species that would just have arisen within a specific ancestral host species colonized another host species. Transfers usually take place when the new host species (receiver), is either phylogenetically or ecologically closely related to the former host species (donor). Finally, extinction that refers to the loss of parasite species is equivalent to what happens within free-living organisms. Though extinction may sometimes be difficult to demonstrate because of parasite sampling bias within host species [5], it may be viewed in two ways. Firstly, parasite extinction due to the speciation of an ancestral host species whereby only one of the daughter host species preserves the ancestral parasite species. This process has been described as “Missing the Boat” [5]. Secondly, following the speciation of the ancestral host species, both daughter host species preserves the ancestral parasite species, but one of the parasite species goes extinct. This process has been described as “Drowning on Arrival” [5]. Whichever one of the two processes is involved in parasite extinction, it is still very difficult to derive firm conclusions.
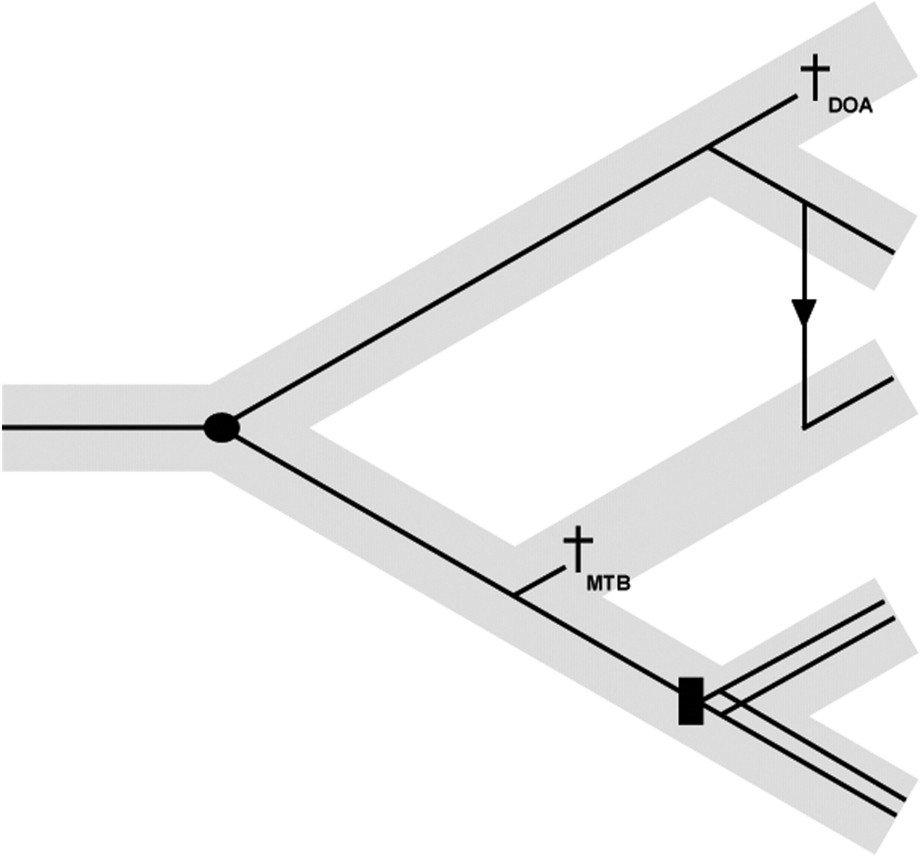
Evolutionary events describing the processes of parasite diversification. The grey and black narrow lines correspond respectively to host and parasite relationships. The black circle refers to cospeciation, the black rectangle to duplication, the arrow to host-switching and crosses to extinction (MTB means “Missing the boat” and DOA “Drowning on Arrival” [5]).
2 Cospeciation: Myth or reality?
One of the main challenges in cophylogeny is to infer processes of parasite diversification over host evolution. In the lack of invertebrate fossil remains, evidence of cospeciation events allows correlations of parasite divergences to geological times, which are given from the host paleontological record. Ultimately it helps to assess the relative molecular evolutionary rates betweens groups of very divergent organisms. So far, several studies were conducted to track the cophylogenetic history within host and parasite assemblages that encompass a large diversity of parasitic organisms among which viruses, bacteria, protozoans, crustaceans, insects and platyhelminths [2,3,10–30]. Surprisingly, cospeciation was not a widespread process which may explain the evolution of parasites across their definitive hosts. The only proofs of codivergence were revealed within viruses, bacteria, protozoans and lice within invertebrate and vertebrate hosts [11,12,14,19,29–36]. Paterson and Banks [4] reported that parasites with greater opportunities of vertical transmission were more likely to cospeciate with their hosts than organisms with horizontal transmission, as it was illustrated within lice and their vertebrate hosts. Conversely, cophylogenetic studies that were conducted within organisms with mainly horizontal transmission, as it is the case with the majority of platyhelminths, showed on the whole different patterns of evolution. Fish parasites that belong to the class Monogenea of the phylum Platyhelminthes would have preferentially diversified through repeated host switching events, as it has been exemplified within the two genera Lamellodiscus [17] and Gyrodactylus [25], and repeated intra host duplication events such as for instance within Dactylogyrus [23]. The proximity of numerous distinct host fish species in aquatic environment and the diversity of microhabitats within branchial cavities could have enhanced these global patterns. Light and Hafner [30] expected that cophylogenetic studies, involving organisms other than the extensively studied pocket gophers (Rodentia: Geomyidae) and their chewing lice (Phthiraptera: Ischnocera) [3,10,12,29], should bring new insights into the development of both theory and methods of cophylogenetic analysis. We propose that the study of the platyhelminth Polystomatidae that infests preferentially amphibious tetrapods could serve as a novel model to improve cophylogenetic tools as well as to inspect a suite of questions about the evolution of the vertebrate hosts.
3 Systematic and biological background of the Polystomatidae (Platyhelminthes, Monogenea)
Monogeneans are platyhelminth parasites that infest mainly marine and freshwater fishes. Following the classification of Boeger and Kritsky [37], the class Monogenea is divided into two subclasses, the Polyonchoinea and the Heteronchoinea, with the latter being subdivided into two infrasubclasses, the Oligonchoinea and the Polystomatoinea. With the exception of a few species that are classified within the Polyonchoinea [see [38]], all other monogeneans that infest tetrapod hosts are from the infrasubclass Polystomatoinea, family Polystomatidae sensu Sinnappah et al. [39]. This family includes only parasites of amphibious tetrapods such as amphibians, freshwater turtles and the African hippopotamus, as well as one species that was recorded from the Australian lungfish. About 150 polystome species are currently described and are divided into 21 genera of unequal diversity, 16 genera within amphibians, with Polystoma of anuran hosts being the most diversified, three genera within freshwater turtles and two monotypic genera within respectively African hippos and Australian lungfishes. Polystomes are found all over the world except in Polar Regions where turtles and amphibians have never been reported. They mainly differ from all other fish monogeneans by their well-developed opisthaptor with three pairs of cup-like suckers. Sphyranura which is the only genus with only one pair of suckers has been identified as a paedomorphic parasite according to its development and phylogenetic position within the Polystomatidae [39–41]. Polystomes like all other monogeneans are generally host and site specific. Within freshwater turtles, parasites can be found either in the bladder, the pharyngeal cavity or the conjunctival sacs (Fig. 2). Hence three different parasite species may live on the same chelonian host species and even in the same individual. Within amphibians with the exception of some salamanders, polystomes (when mature) are found in the urinary bladder (Fig. 3). However, at the juvenile stage, they may be found on the gills of tadpoles, in the kidneys or in the Mullerian ducts. Lastly, they are reported from the skin of a few species of salamanders and the Australian lungfish, as well as the conjunctival sacs of the African hippopotamus.
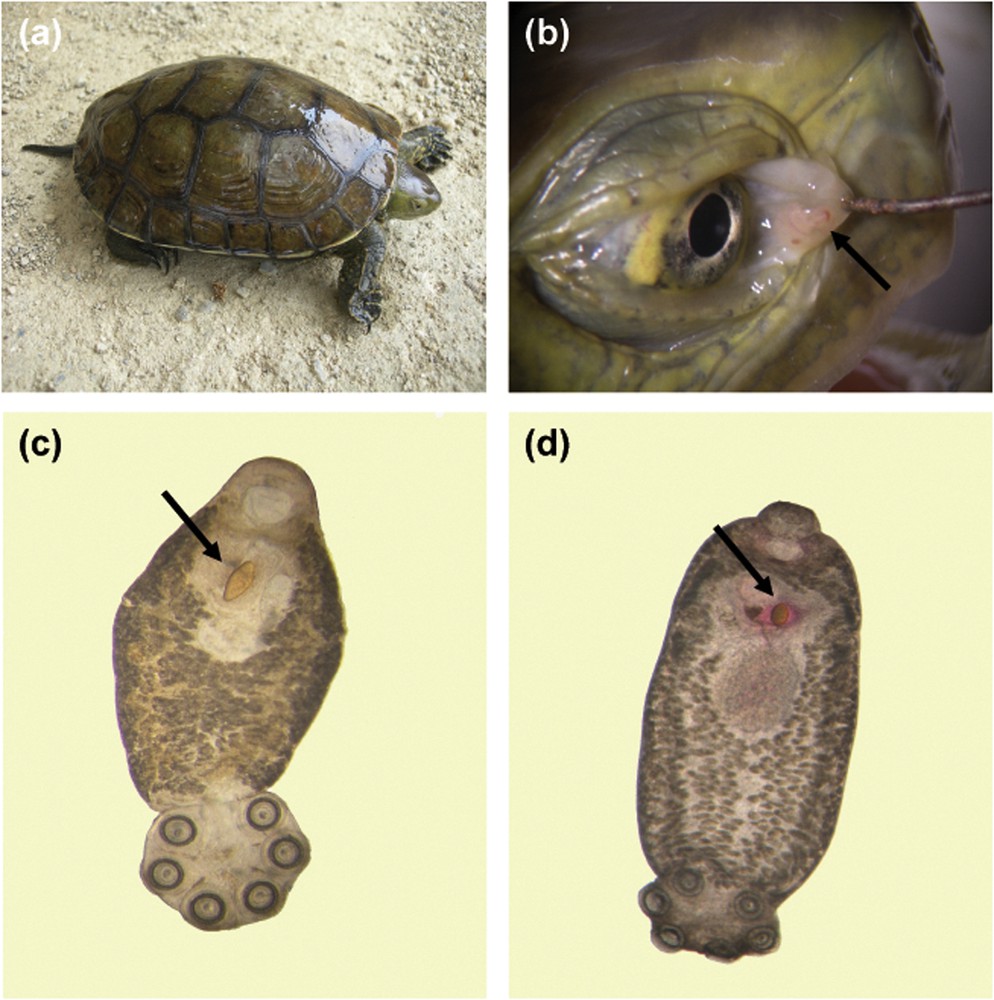
Chelonian host and polystomes. (a) The Mediterranean turtle, i.e. Mauremys leprosa, an indigenous freshwater turtle surveyed in the South of France near Banyuls. (b) Two polystomes under the nictitating membrane of M. leprosa. (c) Eye polystome, i.e. Neopolystoma sp., of M. leprosa. The arrow indicates the location of the diamond shaped egg that characterizes eye polystomes. (d) Bladder polystome, i.e. Polystomoides sp., from the European pond turtle, i.e. Emys orbicularis. The arrow indicates the location of the pear shaped egg that characterizes bladder and pharyngeal polystomes.
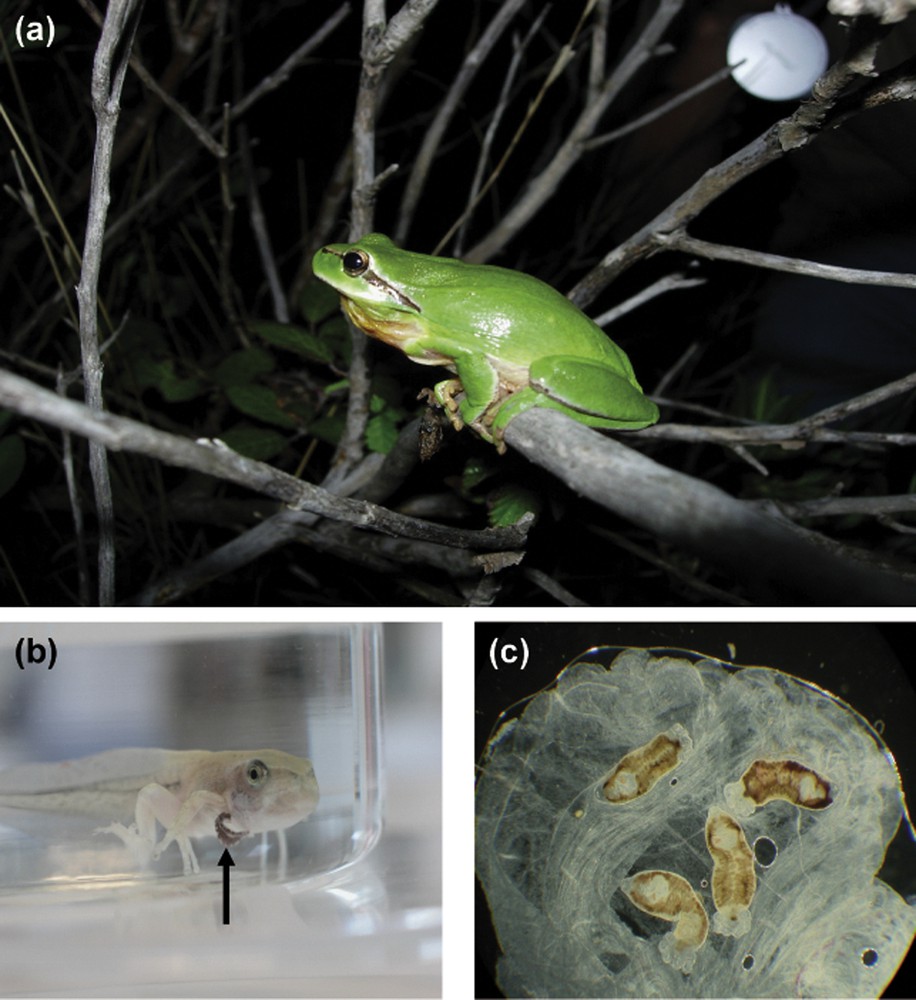
Amphibian hosts and their polystomes. (a) The Stripeless Tree Frog, i.e. Hyla meridionalis, collected in the South of France near Opoul. (b) Polystome hanging from the branchial cavity of a H. meridionalis tadpole. (c) Bladder polystomes of H. meridionalis.
Polystomes retain the typical direct life cycle of monogeneans involving one single host species. Adult worms lay eggs in water that hatch usually within three-four weeks. Oncomiracidia which are free swimming larvae attach to the skin of a new host and after migration colonize the specific ecological niche where they develop into mature forms (Fig. 4a). Different strategies of transmission were selected among polystomes to maximize transmission efficiency. For instance Pseudodiplorchis americanus, parasitic in the desert couch's spadefoot toad Scaphiopus couchii, experiences ovoviviparity and deposits fully developed eggs that hatch immediately during the short reproduction period of its host [42] (Fig. 4b). Eupolystoma that mostly infests the African Amietophrynus has an internal life cycle that may increase exponentially the number of parasite worms within the same host individual. Like for P. americanus, transmission from host to host takes place during host mating [43,44]. Some species of the Polystoma genus, which are mostly specific parasites of neobatrachian hosts, are able to complete their life cycle either on the gills of tadpoles or inside the bladder of adult frogs [45–54] (Fig. 4c). When larvae attach to a young tadpole, they develop rapidly, reaching maturity about three weeks later, reproducing and dying during host metamorphosis [46,55,56]. On the other hand, when they attach to an older tadpole, they develop slowly, migrating to the bladder during host metamorphosis and reaching maturity one to three years later when its host reproduces for the first time.
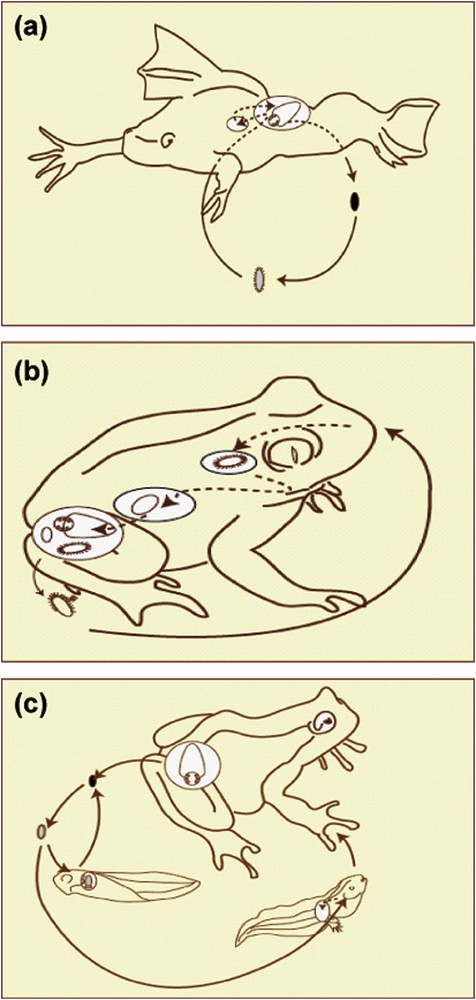
Typical life cycles of amphibian polystomes. (a) Direct infestation – rear entry. Found in Protopolystoma, Eupolystoma, occasionally Polystoma and chelonian polystomes. (b) Direct infestation – anterior entry. Displayed by Pseudodiplorchis. (c) Indirect infestation – rear entry. The typical Polystoma life cycle with a long cycle involving a developmental phase in the branchial chamber of the host tadpole.
4 Evolutionary hypotheses on an early origin of the Polystomatidae
Rohde and Pearson [57] suggested that the present distribution of chelonian polystomes may reflect an ancient distribution going back to at least 200 million years (My), before the break-up of Pangaea. Sinnappah et al. [39] concluded from phylogenetic evidences that the ancestor of the turtle polystome lineage could be even more ancient than 200 My. Similarly Prudhoe and Bray [58] suggested that Polystoma was already distributed among anurans in the Early Cretaceous. Based on biological features of polystomes including the life cycle of anuran polystomes, host specificity, host spectrum, and worldwide distribution, it was assumed that the origin of the Polystomatidae could be very early [59]. Considering also that the paedomorphic polystome Sphyranura oligorchis could be the missing link between the fish monogenean parasites and the branchial adult forms that reproduce on young tadpoles [39] (see Figs. 3b and 4c) and that lungfishes, which are the sister taxon of tetrapods, are the only fishes to be infected by a polystome, it could be hypothesized that the origin of the Polystomatidae dates back to the ecological transition between aquatic and land vertebrates [20].
5 Findings of the evolutionary history of the Polystomatidae
In a phylogenetic framework where phylogenetic relationships of the main genera of the Polystomatidae family, i.e. Concinnocotyla, Neopolystoma, Polystomoides, Sphyranura, Protopolystoma, Pseudodiplorchis, Neodiplorchis, Eupolystoma and Polystoma, were investigated from partial 18S rDNA sequences, Verneau et al. [20] illustrated several key events throughout the polystome evolution that were intimately linked to the host evolutionary history (Fig. 5). Concinnocotyla australensis, which is the lungfish's parasite, was the first polystome to diverge within the Polystomatidae and both turtle and amphibian polystome lineages were monophyletic as well as sister groups. Within amphibian polystomes S. oligorchis, the parasite of the salamander Necturus maculosus, was nested within anuran polystomes but its relationship with neobatrachian and archaeobatrachian polystomes was unresolved. Archaeobatrachian polystomes did not form a clade while neobatrachian polystomes were monophyletic as are indeed their respective anuran host species [60–63]. The basal position of the Australian lungfish's parasite within the Polystomatidae as well as the global phylogenetic arrangements within polystomes evidenced a very ancient origin for the family that may track the evolutionary history of the first aquatic tetrapods following the Actinopterygii–Sarcopterygii transition in the Palaeozoic age, about 425 Million years ago (Mya). Subsequently the origin of both ancestral turtle and amphibian polystome lineages would be related to the split between lissamphibians and amniotes in the Lower Carboniferous, about 350–355 Mya, according to molecular dating that were attempted to calibrate the main cladogenetic events. Early diversification of chelonian and lissamphibian polystome lineages could be correlated to the origin and diversification of their host species groups, respectively 208 and 250 Mya. Concerning the last dating, molecular calibrations we inferred from analysis of combined complete 18S and partial 28S rDNA sequences (Badets and Verneau, unpublished results) showed that the early diversification of lissamphibian polystomes was closer to 300 My than to 250 My, which is more consistent with recent molecular dating assessed within amphibian hosts [61,62]. Consequently, Verneau et al. [20] concluded that the polystomes' evolution was intimately related to the evolutionary line of their host lineages over hundreds of million years.
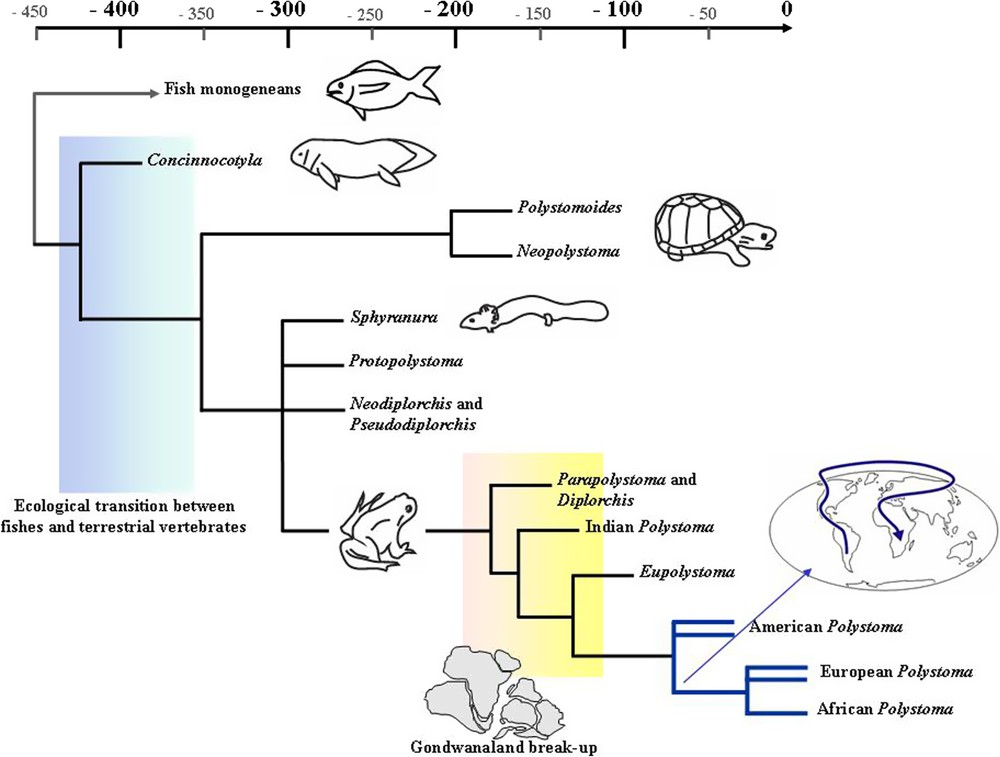
Schematic reconstruction of the evolutionary relationships of the Polystomatidae. Redrawn from [20,64–66].
At smaller time scales, Bentz et al. [64] showed from analyses of ITS1 and partial 18S rDNA sequences that the historical biogeography of Polystoma, the most diversified genus within the Polystomatidae, was as a whole illuminated by the evolution of their host species, namely neobatrachian frogs. Evolution and dispersal of Polystoma from South America to North America on one hand and from North America to Eurasia on the other would be intimately related to the dispersal of either ancestral bufonids or hylids in Paleocene times and by the mid-Cainozoic respectively (Fig. 5). Those results were consistent with previous ITS1 phylogenetic analyses that were conducted within European and African Polystoma species [65]. The paraphyly of European Polystoma species with respect to the monophyly of African ones suggested a single event of colonisation of ancestral European polystomes to Africa. All these results together led Bentz et al. [64] to conclude that if the historical biogeography of polystomes was illuminated by the evolution of their hosts, reciprocal illumination in this host-parasite association may help in inferring the evolution and dispersal of neobatrachians from their origins in the Mesozoic period to recent geological times.
Because neobatrachian polystomes are widely distributed across almost all continents and subcontinents, a Gondwanaland origin could be assumed for the ancestral lineage. Badets et al. [66] investigated the phylogenetic relationships within neobatrachian polystomes that were sampled from Australia, Africa, South and North America, Eurasia and India. Polystome and amphibian trees were inferred respectively from the analysis of concatenated nuclear 18S and partial 28S rDNA sequences and mitochondrial 12S and 16S rDNA sequences. It was shown from cophylogenetic analyses that widespread cospeciation did not occur across the evolution of neobatrachians and their specific polystomes. On the other hand, whenever host switching and duplication events were suggested to explain discordances between host and parasite phylogenies, they were in total contradiction with the overall distribution of polystomes as well as with the main conclusions of Bentz et al. [65] about the origin and evolution of African Polystoma. Therefore, biogeographic vicariance analyses supplemented by molecular calibrations were conducted to investigate the historical biogeography of neobatrachian polystomes. Results showed that the four polystome lineages may be ascribed to centres of diversity, namely Australia, India, Africa and South America and that their relationships substantiated by molecular dating may reflect sequential origins during the break-up of Gondwanaland in the Mesozoic period. Neobatrachian polystome lineages would reveal rifting and drifting of ancient and present continents and to a lesser extent codivergences between hosts and their parasites. Because polystomes show a direct life cycle that involves a short free-living aquatic larval stage, it is likely they are disseminated only passively by their hosts. Thus, the evolutionary pathways of polystomes can bring new fundamental insights about the historical biogeography of their specific hosts, even if little cospeciation has occurred.
6 Prospects
Accordingly, the congruence between vertebrate and polystome phylogenetic branching patterns at deeper levels of times than so far explored within monogeneans, as well as the correspondence between phylogenetic relationships of neobatrachian polystomes and plate tectonics in the Middle Jurassic, provides an exceptional temporal framework for estimating molecular substitution rates within the Polystomatidae. In turn, it may supply a valuable molecular clock to gauge molecular calibrations in fish monogenean parasites that lack widespread cospeciation over host evolution.
It has been shown that polystomes provide unique evidences of historical dispersal events of their neobatrachian hosts, even in the absence of widespread codivergence. Since we discovered polystomes within endemic Madagascan amphibian species (Du Preez, Vences and Verneau, unpublished observations), their phylogenetic relationships will be investigated in order to infer the origins of both host and parasite lineages in Madagascar: Did they colonize the island by overseas rafting or did they originate in Madagascar next to the Gondwanaland break-up?
For several decades, numerous species were dispersed by humans in their non-native home ranges. This was the case for the invasive red-eared slider turtle Trachemys scripta elegans which was introduced as a pets since the 1970s in France and which now coexists with two indigenous endangered freshwater turtle species, the European pond turtle (Emys orbicularis) and the Mediterranean turtle (Mauremys leprosa). All these turtles will be surveyed for their polystomes in natural environments as well as in turtle farms where individuals occur together in order to investigate parasite communities and the hazardous effects of parasite transfers if any.
At last, the plasticity of biological life cycles observed within Polystoma species led us to consider that polystome adults that reproduce on tadpoles may remind the ancestral forms of polystomes, as monogeneans are originally fish parasites. We plan to investigate the developmental mechanisms that are involved in the adaptative plasticity of both life cycles in order to explore the former origins of parasitism.
Acknowledgements
We are indebted to Hervé Le Guyader and Claude Combes for their advice during the writing of the manuscript and to Sophie Bentz for polystome life cycles drawings. O.V. and L.D.P. acknowledge the support of the CNRS and the South African National Research Foundation.


