1 Introduction
Land plants are fixed organisms which have to integrate multiple endogenous and environmental factors to coordinate the differentiation and development of their different parts according the environmental conditions. This coordination is based on the action of small signaling molecules which act at very low concentrations and provide a means of communication between cells and organs. The ‘classical’ phytohormones, identified during the first half of the twentieth century, are auxin, abscisic acid, cytokinin, gibberellin and ethylene. More recently, several additional compounds have been recognized as hormones, including brassinosteroids, jasmonate, salicylic acid and nitric oxide [1]. It is very likely that others have to be identified with a more specific role in the different processes of plant development.
In plants, during post-embryonic development, meristems are initiated at the axils of leaves that will develop a few nodes (short internodes and leaves) to give axillary buds. Many factors will influence the fate of each bud, such as nutrition, photoperiod, plant density (quality of light), or position of the axillary bud itself along the main stem. For each axillary bud, integration of these factors will result in its outgrowth to give a branch or to its maintenance in a dormant state. Recently, the study of high-branching mutants in several species has demonstrated the existence of a novel, graft-transmissible signal, which acts as a branching inhibitor. This work has culminated in the recent identification of this branching inhibitor as a strigolactone, or derived compound which suppresses the outgrowth of a bud located at the axil of leaves [2,3].
2 The classical theory of apical dominance
Before the use of developmental mutants, early studies of shoot branching in the 1930s focussed on decapitation-induced bud outgrowth [4]. The pea was one of the first systems used to study apical dominance. During the vegetative phase, most pea cultivars present dormant axillary buds at most nodes and axillary buds separated by long internodes make such studies easy. After decapitation, axillary buds that were dormant enlarge and start to grow. The term ‘apical dominance’ was used because the removal of the shoot apex leads to the release of dormant axillary buds to form branches. Auxin application studies provided evidence that the hormone auxin, produced in the shoot tip, was involved in the inhibition of axillary bud outgrowth at nodes below [4]. The development of these new stems, also activated by cytokinin produced in roots, allows the plant to survive and to reproduce. The presence of dormant axillary buds at the axil of each leaf and the mechanism of apical dominance have probably evolved in plants as a survival response to herbivory. For several years, auxin and cytokinin were known as the only two hormones controlling branching through apical dominance. Further studies led to the hypothesis that auxin required a second messenger, to inhibit branching [5]. In particular, the two-shoot experiments from Snow [5] suggested that this unknown long-distance signal was moving in the plant with a root-to-shoot direction very likely in the xylem. These “two-shoot” plants were obtained by decapitation of young pea or Vicia faba seedlings to get two similar cotyledonary shoots. When only one cotyledonary shoot was decapitated, the axillary buds on this shoot were inhibited by the apex of the second intact shoot.
More than a decade ago, the detailed physiological characterization (grafting, hormone quantifications) of the high-branching rms pea mutants, demontrated the existence of a novel signal, in addition to auxin and cytokinin, which represses axillary bud outgrowth.
3 Existence of a novel carotenoid derived branching inhibitor
3.1 The genetic approach
In several species, screening for high branching (tillering in rice) mutants led to the identification of the more axillary growth (max) mutants in Arabidopsis, the ramosus (rms) mutants in pea (Fig. 1), the dwarf (d) mutants in rice and the decreased apical dominance (dad) mutants in Petunia (Petunia hybrida) [6–9]. The mutations are all recessive and relatively non pleiotropic. The other characters affected are the stem width and the height of the plant, particularly in rice where the mutants were called dwarf (d) or high tillering dwarf (htd) mutants. These other effects were supposed to be linked to the high branching of the plant. In Arabidopsis, 4 MAX genes were identified and 5 RMS genes in pea.

Pea rms1 high branching mutant plant (right) and wild-type plant (left).
3.2 Grafting experiments with the ramosus (rms) high branching mutants from the pea
Grafting has long been used to study long-distance transport and signaling between different plant tissues. In the pea, epicotyl wedge grafting using 7-day-old plants is very easy to perform. When the rms1 (or rms5) mutant shoot is grafted on a wild-type rootstock, branching in the scion is repressed (Table 1). This result suggested the existence of a graft-transmissible signal produced in WT rootstock able to repress bud outgrowth in mutant scion. The reciprocal graft demonstrated that the signal is also produced in shoots, because WT shoots did not branch when grafted to mutant rootstocks [10]. Whereas branching is inhibited in shoots of reciprocally grafted mutant and WT plants, it is not inhibited in reciprocal grafts between rms1 and rms5 (Table 1). This result suggested that the RMS1 and RMS5 genes control the biosynthesis of the same graft-transmissible signal and that they have to act in the same tissue [11]. In contrast, when rms3 or rms4 mutant shoot is grafted on a WT rootstock, branching is not inhibited (Table 1). These mutants could be considered as response mutants [6]. More complex two-shoot grafts with WT and rms1 shoots growing from the same rms1 rootstock and showing different branching phenotypes provided evidence that the signal was able to move only acropetally in the shoot (with a root-to-shoot direction) probably through the xylem [12]. Similar grafting studies have been reported in Arabidopsis [13,14] and petunia [9].
Phenotypes of the different graft combinations between WT and three rms pea mutants.
| Genotype of rootstock | Scion | WT | rms1 | rms5 | rms4 |
| WT | Non Br | Non Br | Non Br | Br | |
| rms1 (ccd8) | Non Br | Br | Br | Br | |
| rms5 (ccd7) | Non Br | Br | Br | Br | |
| rms4 (F-Box) | Non Br | Non Br | Non Br | Br |
All these studies led to a new model on the control of branching in the pea [6,15]. In this model, the RMS1 and RMS5 genes control the biosynthesis of a novel branching inhibitor while the RMS3 and RMS4 genes are involved in the response to the novel signal. The cloning of these genes supported the model and suggested that the signal was carotenoid derived.
3.3 Cloning of the branching genes
The first branching genes cloned were the MAX genes in the model plant Arabidopsis thaliana [14,16–18]. They were shown to have their homologs among the branching genes in other species (Table 2) which indicates that the genetic network is conserved between dicotyledons and monocotyledons. Recently, rice has proven to be an important model plant as two novel branching genes have been cloned in this species [19–22].
Genes controlling branching in Arabidopsis, pea and rice.
| Arabidopsis | Pea | Rice | References | |
| Encoded protein: | ||||
| Biosynthesis | ||||
| CCD7 | MAX3 | RMS5 | D17/HTD1 | [14,23,53] |
| CCD8 | MAX4 | RMS1 | D10 | [16,54] |
| Iron containing protein | D27 | [19] | ||
| Cytochrome P450 | MAX1 | [17] | ||
| Signaling | ||||
| F-Box | MAX2 | RMS4 | D3 | [18,23,26] |
| Hydrolase | RMS3 ? | D14/D88/HTD2 | [20–22] | |
| TCP | BRC1/BRC2 | PsTB1 | FC1/OsTB1 | [55,56] unpublished |
3.3.1 The biosynthesis genes
Consistent with the grafting experiments, RMS1 and RMS5 encode two enzymes, two Carotenoid Cleavage Dioxygenase (CCD8 and CCD7, respectively) which are both targeted to the plastid [16,23]. These genes belong to a family containing nine genes in Arabidopsis, which encode enzymes known to cleave carotenoids. In Arabidopsis, five genes of this family are involved in the biosynthesis of abscisic acid (ABA), a well-known plant hormone originating from the cleavage of epoxy-carotenoids [24]. The exact substrates and functions of CCD7 and CCD8 in plants are not known, but it has been proposed that the branching inhibitor could be biosynthesized from carotenoids [25]; this information has been an essential clue to its discovery (see below).
MAX1 from Arabidopsis encodes another enzyme which would act downstream of MAX4 (CCD8) and MAX3 (CCD7) [17]. MAX1 belongs to the large family of cytochrome P450 s, many of which are involved in the biosynthesis of other plant hormones. Recently, the rice tillering gene D27 has been shown to encode a novel plastid targeted enzyme, an iron-containing protein with no homology with any functionally identified protein [19].
3.3.2 Genes involved in the response/signaling pathway of the branching inhibitor
MAX2, RMS4 and D3 are orthologous members of the F-box leucine-rich repeat (LRR) protein family (Table 2). This result supported the hypothesis that they have a role in the response/signal transduction of the novel hormone [18,23,26] and suggested that the signaling pathway of the branching inhibitor has similarities with the signaling pathways of other major plant hormones (auxin, gibberellic acid, jasmonic acid). These pathways involve ubiquitination of specific targeted proteins by a Skp/Cullin/F-box complex (SCFMAX2) and their subsequent degradation by the 26S proteasome [27]. Recently, the rice D14/D88/HTD2 gene has been shown to encode a member of the α/β-fold hydrolase superfamily [20–22] in which proteins share a similar 3D-structure [28] and which contains GID1, a gibberellin receptor [29].
In maize, the transcription factor TEOSINTE BRANCHED1 (TB1) has long been known to suppress axillary bud outgrowth in maize and to be one of the few genes involved in maize domestication from its wild and highly branched ancestor, teosinte [30,31]. In comparison to teosinte, TB1 is upregulated in maize axillary meristems [31]. In Arabidopsis, mutants of the genes BRC1 and BRC2 have a highly branched phenotype and the encoded proteins are members of the TCP plant-specific family of transcription factors (the TCP family is named after TEOSINTE BRANCHED1 (TB1) in maize, CYCLOIDEA (CYC) in Anthirrinum majus, and PCF in rice), involved in multiple developmental control pathways [32].
4 Strigolactones (SL) as the novel branching inhibitor
4.1 Roles of SL in the rhizosphere
SL are a group of small molecules produced by the roots of plants and exuded in the rhizosphere where they play major roles in both parasitic and symbiotic interactions [33,34]. Since 1973, seed germination of the parasitic weeds Orobanche and Striga has been known to be stimulated by SL in the soil where they signal the presence of a host plant [35]. These parasitic plants, producing numerous and extremely small seeds, are major agriculture pests in Africa (Striga) and in more temperate regions around the world (Orobanche) [36]. The same class of compounds has recently been shown to play a major role as root recognition signals in the symbiotic association between plants and arbuscular mycorrhizal (AM) fungi [37,38]. In the arbuscular mycorrhizal symbiosis, plants obtain water and mineral nutrients from their fungal partners, enabling them to survive under various stressful conditions. Arbuscular mycorrhizal fungi are obligate symbionts and depend on the carbon provided by their plant host to achieve their developmental cycle. Strigolactones are also produced by non-mycotrophic plants, such as Brassicaceae (including Arabidopsis) which, consistent with what is now known of their role in growth regulation, indicated that they may have additional functions in plants [39]. In 2005, Matusova et al. showed that strigolactones are carotenoid derived [40]. Therefore, strigolactones or related compounds became good candidates to correspond to the branching inhibitor.
4.2 SL, the branching inhibitor
To identify the genes involved in SL biosynthesis, researchers working on root colonization by AM fungi or parasitic seed germination looked for mutants in Carotenoid Cleavage Dioxygenase (CCD) genes. One of the first important results obtained with the rms pea mutants was that root exudates of rms1 (mutated in the pea CCD8 homolog) failed to stimulate proliferation of fungal hyphae or Orobanche seed germination when compared to WT exudates. In contrast, rms4 which exhibits the same phenotype of profuse branching but was thought not to be defective to the branching inhibitor synthesis but to its signal transduction, was shown to produce normal or even higher levels of strigolactones [2]. These results strongly suggested that CCD8/RMS1 (and CCD7/RMS5) are necessary for the synthesis of strigolactones. Another essential result was the use of GR24, a synthetic analog of SL. Direct application of the synthetic strigolactone GR24 on the axillary bud of rms1 mutant plant is able to inhibit its outgrowth at a concentration of 100 nM. Using a method to feed GR24 to the vascular stream of pea shoots, ten nanomolar GR24 was sufficient to inhibit bud outgrowth at the two nodes above the feeding site (but not at nodes below). No inhibition was observed when treating with GR24 the rms4 response mutant [2]. Consequently, strigolactones have all the characteristics of a plant hormone: action at very low concentrations, possibly at long distance, complementation of phenotype of the biosynthesis mutant by exogenous applications and no complementation with the response mutant.
That a series of well defined pea branching mutants was available and that axillary buds are easily accessible to exogenous treatments and to precise observations contributed to this discovery. At the same time, Umehara et al. used a series of high tillering rice mutants to demonstrate the implication of SLs in tillering in monocots [3].
4.3 Natural strigolactones
Strigolactones present the same basic skeleton of an ABC tricyclic lactone connected via an enol ether bridge to a butyrolactone D ring (Fig. 2). The CD part of the molecule is thought to be the essential part for bioactivity as germination stimulants of parasitic weeds [41]. Today, about 15 natural strigolactones have been identified in root exudates of monocotyledonous and dicotyledonous plant species [33], strigol and strigyl acetate being the first to be identified in cotton root exudates in 1966. The identification and structural characterization of strigolactones is particularly difficult because they are produced by plants in extremely low quantities and are relatively unstable.
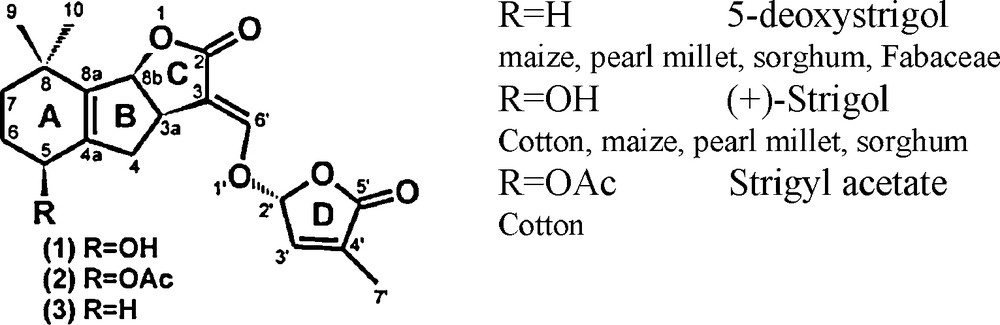
Chemical structures of some natural strigolactones.
4.4 Ecological importance of SL for early colonization of land by plants
Genes involved in the biosynthesis (CCD7 and CCD8) of strigolactones can be found in the genome of tree species, such as poplar or apple or in non-vascular organisms, as the Bryophyte Physcomitrella patens in which strigolactones have been identified in tissues (Yoneyama, pers. comm.). It is very likely that these compounds are involved in the control of the architecture of most land plants. Molecular data and studies of fossils suggest that arbuscular mycorrhizal symbiosis is very ancient (more than 400 million years) and has facilitated the adaptation and evolution of primitive plant species to life on land [42]. All this suggests that strigolactones that were selected to facilitate the process of host recognition throughout the plant kingdom have played a major role in the colonisation of land by plants and have facilitated the development of large vegetative shoot among vascular plants.
5 Regulation of SL biosynthesis
SL biosynthesis in roots and exudation in the rhizosphere have been shown to be regulated by nutrient availability in the soil, in particular phosphorus (P) and nitrogen (N). In red clover, a reduced supply of P significantly increases orobanchol exudation from roots [43]. In sorghum, not only P deficiency but also N deficiency markedly promoted 5-deoxystrigol exudation and SL content in roots [44]. These results indicated that plants respond to phosphorus and nitrogen depletion in the soil by enhancing exudation of SL; presumably this has been selected because it acts as a signal for AM fungi which are known to supply mineral nutrients, particularly P, to host plants. In the same study [44], no change in shoot content of 5-deoxystrigol was observed.
In studies on the control of branching, regulation of SL biosynthesis has been investigated with the analysis of transcriptional regulation of biosynthesis genes. In the pea, the RMS1/CCD8 gene has been shown to be finely regulated by auxin and by an auxin-independent feedback signal (most studies have been done using epicotyl tissue). After shoot decapitation, there is a massive and rapid drop in the RMS1 transcript level which is restored if auxin is applied to the decapitated stump [45]. This result suggests that auxin positively regulates the RMS1 transcript level, which could be the mechanism by which auxin regulates shoot branching in apical dominance; hence SL would be the secondary messenger to auxin in apical dominance [45,46]. Auxin has been shown also to regulate cytokinin levels in root xylem sap and in the nodal stem [47,48]. Consequently, auxin can suppress bud outgrowth in apical dominance by controlling both SL and cytokinin biosynthesis. How SL and cytokinin interact to control bud outgrowth is still unknown and is a matter of future research. In four out of five rms high-branching mutants, RMS1 is strongly up-regulated to levels that cannot be achieved by auxin applications. These mutants do not have elevated auxin levels which indicates the existence of an auxin-independent feed-back signal, regulating SL biosynthesis in the stem [45]. This shoot-to-root graft-transmissible signal, has also been shown to regulate cytokinin concentrations in root xylem sap [49,50].
6 Perspectives
Another theory has been proposed on how SL could control bud outgrowth based on competition for auxin transport in the main stem between apices. This competition would result from the limited capacity of the main stem to transport auxin [51] or, based on the canalization hypothesis developed by Sachs in 1970, from a positive feedback between auxin flux and polarization of auxin transport [52]. How this theory could be reconciled with a localised action of SL in the axillary bud is a major question for future research.
The discovery that strigolactones form a novel class of plant hormones opens a new field of research in plant biology. It is very likely that other roles of SL in plant development will be discovered in the near future. New enzymes in the SL biosynthesis pathway have to be identified to get the tetracyclic SL molecule from a carotenoid substrate. The signaling pathway should present similarities with the signalling pathways of other hormones such as auxin, gibberellin or jasmonic acid where proteins are targeted for degradation via the SCF complex. It is well known that one important factor controlling shoot branching is the availability of nutrients such as nitrogen. The extent to which strigolactones are a signal which relays the state of nutrients in soil to the shoot and controls growth by branching is another important aspect for future research.


