1 Introduction
The actual productivity of crops usually falls far short of its maximum potential, mainly due to various environmental constraints, such as drought, extremes in temperature, salinity, etc. Such constraints are expected to augment in coming decades due to global warming and associated changes in climate. Thus, to sustain the present rate of increase in agriculture production, it is important to develop improved varieties of crop plants with higher tolerance to various stresses. Better understanding of the mechanisms by which a plant can cope with adverse environments is important for the development of stress-tolerant plants. Studies in various plant species, especially in model plants such as Arabidopsis and tobacco have revealed various mechanisms involved in stress tolerance of plants, and those mechanisms were summarized in various reviews [1–3]. To make use of accumulating information on stress tolerance mechanisms of plants for the development of stress-tolerant varieties of a particular agricultural crop, it is important to investigate the specific stress tolerance mechanisms of that particular crop too. Therefore, it is time for us to strengthen our understanding of the stress tolerance mechanisms working in agriculturally important plants, such as potato, tomato, rice, etc.
Potato (Solanum tuberosum L.) is highly sensitive to environmental stress. The effect of environmental constraints on potato cultivation is well reflected in the discrepancy between its average and record yields; according to Boyer [4], the maximum yield of potato is three times larger than its average yield, and this difference is mainly due to environmental constraints. Potato plants are very sensitive to drought stress [5]; hence, drought is a major constraint for potato cultivation in many parts of the world [6,7]. The adverse effect of drought stress on potato virtually occurs at all stages of the crop, from seedling emergence to tuber initiation and bulking, which ultimately results in reduced tuber yield [8,9]. Furthermore, prolonged water scarcity leads to several physiological disorders, such as tuber cracking, tuber malformation, hollow heart, vascular discoloration and reduction in the accumulation of total dry matter in tubers [10,11]. Overall, the effect of drought stress on potato cultivation is severe.
In future, it will not be economical to irrigate the increasing proportion of drought-struck agricultural land, and hence, the development and supply of drought-tolerant varieties of potato is increasingly important. There is also an urgent need to supply improved varieties to farmers so that they will continue potato cultivation in hot and dry areas. Faster and better understanding of drought-tolerance mechanisms in potato will help us to speed up crop improvement programs (through breeding and/or genetic engineering) for the development of drought-tolerant verities. Even though there were few studies to deduce the drought-tolerance mechanisms in potato, an understanding of the key genes and overall network of genes that are related to drought stress in potato is still inadequate.
Recently, a functional screening-based approach, using yeast or bacteria as an experimental system, is being utilized to identify genes that may play significant roles in the stress tolerance of plants [12–17]. Among them, a functional screening system that uses yeast have advantage over those that use bacteria, as yeast is a eukaryotic organism with relatively closer post-translational modification to that of higher plants [18]. Priyanka et al. [19] used a yeast system to screen for multiple abiotic stress tolerance abilities of a pigeon pea hybrid-proline-rich protein, and found that the yeast overexpressing the protein is tolerant to multiple stresses. Functional screening of yeast expressing a cDNA library from Jatropha curcas resulted in the isolation/identification of sequence orthologues of genes with known functions in stress tolerance, such as allene oxide cyclase, late embryogenesis abundant protein-5, metallothionein, thioredoxins, etc.; in addition to these, a number of uncharacterized genes are also identified to have potency to impart stress tolerance [16].
With the availability of a large amount of genome sequence, transcriptome, and proteome data, it is possible to predict potential stress tolerance genes of a crop plant. However, the functional screen-based methods for choosing/identifying potential stress tolerance genes of a crop plant hold their own advantages. These functional screening methods select the genes based on their relative ability in imparting higher stress tolerance to yeast or bacteria; hence, these methods can be extended further to select the most potent genes from a bigger list of potential genes. In addition, these methods are capable of identifying ability of unknown or uncharacterized genes, leading to the discovery of new stress tolerance genes.
In this study, we used a yeast-based functional screen to identify potential drought tolerance genes in potato. To enable the screening process, a cDNA expression library was constructed from drought stressed potato plants under the control of the yeast GAL1 system, and yeast transformants expressing potato cDNAs were selected for their ability to survive in hyperosmotic stress condition. The relative tolerances of the selected yeast transformants to multiple abiotic stresses were also studied to propose the most potent candidate genes for detailed investigations to explore their role in stress tolerance of potato.
2 Materials and methods
2.1 Plant material
Nodal explants of potato were grown in Murashige and Skoog (MS) medium [20], containing 2.5% sucrose and 0.7% agar for 28 days, then the regenerated plants were transferred to a liquid ½MS medium, containing 0.5% sucrose (L½MS) for a week. During these seven days, the culture bottles were fully closed for first two days, partially open for the third and fourth day and fully open for the last three days to make the plants acclimatize to the culture room conditions. During this acclimatization period, at every 24 h the L½MS medium in which plants were growing was replaced with the same fresh medium. Plants were found to remain strong and healthy in the standard culture room conditions (relative humidity of 50–60%, 25 ± 2 °C and a 16 h photoperiod). The plants with similar height and root mass (by visual observations) were used for stress experiments (Fig. 1).

Different stages of in vitro potato plantlet development through nodal culture. Color online.
2.2 Details of yeast strain (Saccharomyces cerevisiae) used for the functional study
Accession No: Y00000, Strain: BY4741, Genotype: MATa; his3 Δ1; leu2Δ0; met15Δ0; ura3Δ0. This strain was provided by the European Saccharomyces cerevisiae Archives for Functional Analysis (EUROSCARF), Frankfurt, Germany.
2.3 Stress treatment of plants and RNA isolation
To give drought stress to the plants, the liquid medium (L½MS) in which the plants were grown during the acclimatization period in the culture room (described above) was replaced with fresh L½MS medium additionally containing 25% polyethylene glycol (PEG 8000, Sigma, St. Louis, USA). After 24 h, the plants were found to show clear signs of wilting (Fig. 2). In the case of the control plants, the L½MS medium in which plants were grown was replaced with fresh L½MS medium; the control plants were found to remain turgid even after 24 h. After 24 h of treatment, the whole plant was removed from the bottle, washed with distilled water and frozen in liquid nitrogen before being stored at –70 °C. Total RNA was isolated from these whole plant samples using the NucleoSpin RNA Plant (Macherey-Nagel, Düren, Germany) kit, following the manufacturer's protocol. The total RNA quantity and quality were estimated using an ND-1000 spectrophotometer (Nanodrop Technologies, Delaware, USA).

Drought stress treatment of plants by providing media containing different concentrations of polyethylene glycol (PEG). Color online.
2.4 Construction of Infusion SMARTer cDNA and its size separation
First-strand cDNA was synthesized from 500 ng of total RNA using the Infusion SMARTer Directional cDNA Library Construction Kit (Clontech, California, USA). Double-stranded cDNA was generated by long-distance PCR amplification of the first-strand cDNA using Accu power HF PCR Pre Mix (Bioneer, Daejeon, Korea). Briefly, 2 μL of single-stranded cDNA were used for PCR amplification and a range of PCR cycles were performed in order to identify the best cycle number. We decided to use 18 cycles of PCR for the synthesis of double-stranded cDNA for further experimentation as per the manufacturer's instructions. The double-stranded cDNA pools were size separated using NucleoSpin columns (CloneTech, California, USA). The yield and quality of the amplicons were monitored both by using agarose gel analysis and ND-1000 spectrophotometer.
2.5 Modification of pYES 2.1/V5-His TOPO vector
In order to facilitate the insertion of Infusion SMARTer cDNA into pYES 2.1/V5-His TOPO vector (Invitrogen, California, USA), the vector was modified by following the instructions given in the Infusion SMARTer directional cDNA library constructions kit (Clontech, California, USA). In short, the vector was amplified by using specially designed forward (PF-5′-TTGATACCACTGCTTAGGGCGAGCTTAATATTCCCT-3′) and reverse (PR-5′-TCTCATCGTACCCCGCTCCTCGGTCTCGATTCTACG-3′) PCR primers, to make it suitable for infusion cloning. Accu power HF PCR Pre Mix (Bioneer Corp, Daejon, Korea) was used for PCR amplification. The PCR product was purified by gel elution and was used for cloning. The modified vector has special insertion sites to enable the infusion cloning of SMARTer cDNA. The modified vector retained all the essential elements of pYES 2.1/V5-His-TOPO vector to act as an Escherichia coli–yeast shuttle vector. The vector can be propagated in E. coli, and the positive transformants can be selected using a bacterial selection marker for Amp r, whereas the transformants in the yeast BY4741 strain (described above) can be selected using a URA3 marker. In addition, cloning of genes of interest downstream of the GAL1 promoter will allow the regulated expression of those genes in yeast, which is the same as that of original pYES 2.1/V5-His-TOPO vector.
2.6 cDNA library construction by infusion cloning of cDNA into the modified pYES 2.1/V5-His TOPO vector and its amplification in E. coli
For cloning cDNA into the vector, Infusion HD cloning kit (Clone Tech, California, USA) was used. As per the instructions given in the kit, 200 ng of purified Infusion SMARTer cDNA and 175 ng of vector were used per the cloning reaction. The product of the cloning reaction was purified using the QuickClean Enzyme Removal Resin (Clontech, California, USA), and was re-suspended in 20 μL of double-distilled water. Two infusion cloning reactions were performed simultaneously, and their products were pooled to obtain a total of 40 μL, from which 5 μL was used per E. coli (E. coli TOP 10F, Invitrogen, Carlsbad, USA) transformation. E. coli transformation was performed using a Gene Pulser Cuvette 0.1 cm (Bio-RAD, California, USA) and a Micropulser electroporation unit (Bio-RAD, California, USA). Each E. coli transformation resulted in approximately 10,000 transformants on the ampicillin selection plates. Eight E. coli transformations were carried out to increase the library titer, each of which were performed with 5 μL of the purified infusion product to result in approximately 80,000 transformants. After 18 h of growth on the selection medium, the transformed colonies were pooled, and the plasmid was isolated using the Gene All Exprep Plasmid Midi prep Kit (Gene All Biotech, Seoul, Korea). Plasmid DNA was quantified using an ND-1000 spectrophotometer.
2.7 Media and growth conditions for the selection of drought-tolerant transformants of yeast
The yeast strain, BY4741 is deficient of de novo uracil production; hence, a synthetic defined medium (control media [CM media], see Table 1), devoid of uracil, was used for the selection of yeast transformants. For the selection of drought stress-tolerant transformants of yeast, a synthetic defined medium containing sorbitol, galactose and raffinose (drought selection media [DSM], see Table 1) was used. To set an optimum concentration of sorbitol in the selection medium for the selection of drought-tolerant yeast transformants, an empty vector was transformed into the yeast strain BY4741 to result in a control yeast cell. A two-day-old single colony of the control yeast cell grown on a CM selection plate was selected and inoculated in 5 mL of liquid CM medium, which was then grown overnight; 100 μL of 10−3 dilution of the culture was plated on DSM media containing various concentrations of sorbitol (1.75 M, 2 M, 2.1 M, and 2.2 M) and was grown for five days. The minimum concentration (Fig. 3) at which the control yeast cells failed to grow was selected to be used for the screening of drought-tolerant yeast transformants. Unless specified, the yeast cultures were grown at 30 °C and for liquid broth cultures, the shaker was set to 250 rotations per minute.
Compositions of the yeast synthetic selection media.
| Name of the medium | Components | Amount per liter |
| Control media (CM) | Yeast synthetic drop-out medium supplemented without uracil (Sigma, Y 1501) | 1.92 g |
| Yeast nitrogen base without amino acids (Sigma, Y 0626) | 6.7 g | |
| For plates, add bacteriological agar (Duchefa, M1002.1000) | 20 g | |
| 20% galactose (Sigma, G0750), filter sterilized (added after autoclaving) | 100 mL | |
| 20% raffinose (Sigma, R7630), filter sterilized (added after autoclaving) |
275 μL | |
| Drought selection media (DSM) | Yeast synthetic drop-out media supplemented without uracil (Sigma, Y 1501) | 1.92 g |
| Yeast nitrogen base without amino acids (Sigma, Y 0626) | 6.7 g | |
| Sorbitol (Sigma, S1876) | Different concentrations (1, 1.5, 1.75, 2, 2.1, 2.2 M) | |
| For plates, add bacteriological agar (Duchefa, M1002.1000) | 20 g | |
| 20% Galactose (Sigma, G0750), filter sterilized (added after autoclaving) | 100 mL | |
| 20% Raffinose (Sigma, R7630), filter sterilized (added after autoclaving) |
275 μL | |
| Salt selection media (SSM) | Yeast synthetic drop-out media supplemented without uracil (Sigma, Y 1501) | 1.92 g |
| Yeast nitrogen base without amino acids (Sigma, Y 0626) | 6.7 g | |
| Sodium chloride (Daejung, 7548-4400) |
1.2 M | |
| For plates, add bacteriological agar (Duchefa, M1002.1000) |
20 g | |
| 20% Galactose (Sigma, G0750), filter sterilized (added after autoclaving) | 100 mL | |
| 20% Raffinose (Sigma, R7630), filter sterilized (added after autoclaving) |
275 μL |
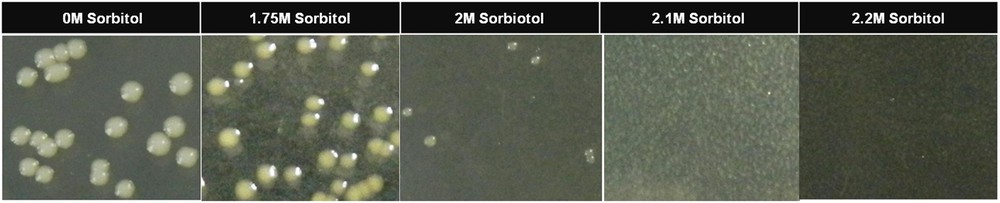
Growth of control yeast cells on drought selection media containing different concentrations of sorbitol. Color online.
2.8 Transformation of potato cDNA library into yeast and screening for drought tolerance
The transformation of yeast was performed by following the method described by Benatuil et al. [21]. Briefly, 300 μL of yeast competent cells were transformed with 5 μL of library plasmid (500 ng of plasmid per μL) by electro-transformation using a Micro Pulser (Bio-Rad, California, USA); the transformation product was grown in half-strength YPD medium (1% yeast extract + 2% peptone + 2% dextrose) containing 0.5 M of sorbitol for 1 h, and the cells were then collected by centrifugation and were re-suspended in 1 mL of the CM medium.
To determine the effect of increasing concentrations of sorbitol on the survival of yeast transformants, we plated 100 μL of the re-suspended transformation products on DSM plates containing varying concentrations of sorbitol (0 M, 1 M, 1.5 M, 1.75 M, 2 M, and 2.1 M), and incubated for five days. According to the observed trend in growth of the transformed cells on these media, DSM plates with a 2.1 M sorbitol medium (2.1 M DSM medium) were used to select for the drought-tolerant yeast transformants (Fig. 4). After five days of incubation, the colonies that had been grown on a 2.1 M DSM medium were picked and streaked on fresh plates containing the 2.1 M DSM medium. To compare the growth of tolerant yeast cells to that of the control yeast cells, the control yeast cells were streaked on each plate, as shown in Fig. 5.
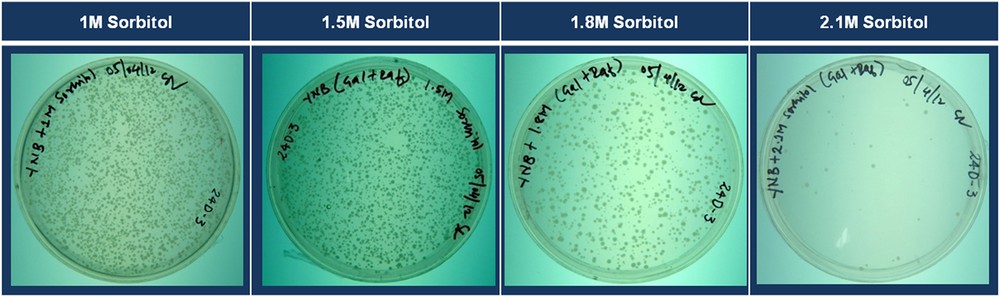
Growth of yeast cells transformed with the potato cDNA library on drought selection media containing different concentrations of sorbitol (the same amount of transformation product was plated on each plate). Color online.
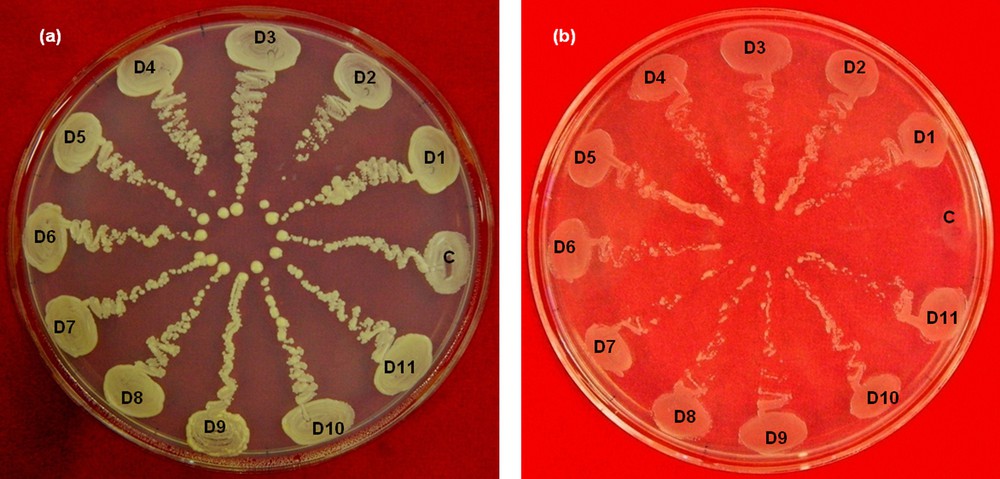
Comparison of the growths of control and tolerant yeast cells. (a) Control plate without stress. (b) Drought selection plate contacting 2.1 mol of sorbitol per liter. Control yeast transformed with empty vector (C), D1 to D11 are drought-resistant transformants of yeast. Color online.
2.9 Determination of the relative tolerance of yeast colonies to drought, salt and high-temperature stresses
To test the relative tolerance of the selected yeast transformants to drought stress, they were grown in the liquid CM medium overnight, and the OD at 600 nm (OD600) was adjusted to 2 in a liquid DSM media containing 2.1 mol of sorbitol per liter. Four dilutions 10−1, 10−2, 10−3, 10−4 of the culture were made in the same liquid medium, 5 μL of each dilution were drop-plated on DSM plates containing 2.1 mol sorbitol per liter, and they were grown for five days. Yeast transformants showing growth at 10−4, 10−3, 10−2 and 10−1 were allocated a relative score of 4, 3, 2 and 1, respectively. The experiment was repeated three times, and the average of the three readings was calculated to compare the relative tolerance of yeast transformants to the stress.
Similarly, to test the relative tolerance of the selected yeast colonies to the saline stress, the OD600 of the overnight grown culture was adjusted to 2 in liquid salt selection media (SSM) containing 1.2 mol of NaCl per liter (see Table 1). Five microliters of different dilutions (10−1, 10−2, 10−3, 10−4) of the culture in the same media were drop-plated onto a plate containing a 1.2 M NaCl SSM medium, and were incubated at 30 °C for five days. A system of scoring as mentioned above was applied.
To study the relative tolerance of the yeast transformants to high temperature, the OD600 value of overnight grown culture was adjusted to 2 in sterile water, and was then serially diluted in sterile water. Five microliters of the different dilutions (10–1, 10–2, 10–3, 10–4) were drop-plated on the CM media (Table 1), and were incubated at 39 °C for three days, and then scoring was again applied in the same manner. The control plate was prepared in the same way, but it was incubated at 30 °C and was observed after three days.
2.10 Plasmid rescue and sequence annotation
Yeast colonies were grown in liquid CM medium for two days, and the plasmid was isolated using the protocol reported by Byrd and St-Arnaud [22]. The plasmid was then transformed into E. coli. Plasmids from positive E. coli colonies (confirmed by colony PCR) were isolated and sequenced at Bioneers pvt LTD, Korea, with vector-specific forward (SeqPF-5′-AATATACCTCTATACTTTAACGTC-3′) and reverse sequencing primers (SeqPR-5′-GATGCGGCCCTCTAGAAACT-3′).
Prior to annotation, the vector portions were removed from the sequences. The sequences were then annotated by using online BLAST programs, available at NCBI http://www.ncbi.nlm.nih.gov/blast/, TIGR plant transcript assemblies http://tigrblast.tigr.org/euk-blast/plantta_blast and in the BLAST search tool available online at the potato genome sequencing consortium project's website, http://solanaceae.plantbiology.msu.edu/integrated_searches.shtml. For the assignment of functional terms to gene products, the Balst2GO application available at www. blast2go.com was used. GenBank accession numbers of all the gene sequences relevant to this article are provided in Table 2, and they are accessible online through the NCBI nucleotide sequence database.
List of the identified genes, their relative ability to enhance tolerance of yeast cells to different abiotic stress and biological process in which the genes are involved.
| Sr. No. | Clone ID | GenBank accession number | Putative function of gene | Relative tolerance of yeast cells expressing the particular cDNA to different stresses | Biological process in which the gene product is involved (based on gene ontology search tool available online, www. blast2go.com) | |||
| Drought | Salt | High temperature | Multiple stress | |||||
| 1 | StDT3 | JX845303 | O-linked GlcNAc transferase | 2 | 1 | 2 | 5 | Glucose catabolic process |
| 2 | StDT10 | JX839744 | Phospholipase D, partial | 2 | 0.7 | 1.7 | 4.4 | Lipid metabolism. Response to cadmium ion. Regulation of stomatal movement. Positive regulation of abscisic acid-mediated signaling pathway |
| 3 | StDT11 | JX839745 | AAA-metalloprotease FtsH, partial | 1 | 0.7 | 2 | 3.7 | Proteolysis |
| 4 | StDT15 | JX683406 | 3-beta-hydroxy-delta-5-steroid dehydrogenase | 2 | 1 | 3 | 6 | Steroid biosynthetic process. Oxidation–reduction process |
| 5 | StDT20, 94, 152, 205 | JX683407 | Non-specific lipid transfer protein 2 | 2 | 1 | 1 | 4 | Lipid transport. Response to stress |
| 6 | StDT21 | JX683408 | N-acetyltransferase | 1.3 | 0 | 2 | 3.3 | Fatty acid biosynthesis. Defense response to insect |
| 7 | StDT22, 122 | JX683409 | Proteinase inhibitor type 2 CEVI57 | 2 | 1 | 2 | 5 | Negative regulation of peptidase activity. Response to oxidative stress |
| 8 | StDT24 | JX683410 | SUMO E2 conjugating enzyme SCE1 | 3 | 1 | 2 | 6 | Protein sumoylation. Embryo development ending in seed dormancy. Response to abscisic acid stimulus |
| 9 | StDT25, 51, 126,166 | JX683417 | Thioredoxin H-type 2 | 1.3 | 1 | 1 | 3.3 | Cell redox homeostasis. Glycerol ether metabolic process. Electron transport chain |
| 10 | StDT26 | JX683411 | Ribosomal protein S27 | 2 | 2 | 2 | 6 | Translation. RNA methylation. Ribosome biogenesis |
| 11 | StDT28 | JX683412 | Cytochrome c | 1 | 0 | 2 | 3 | Electron transport chain/Respiratory chain |
| 12 | StDT29, 150 | JX683437 | Metallothionein-like protein | 2.3 | 1 | 2 | 5.3 | — |
| 13 | StDT30, 93 | JX683428 | Anaerobic basic leucine zipper protein | 1 | 0 | 2 | 3 | Regulation of transcription/Transcription factor. Cellular response to starvation |
| 14 | StDT32, 71 | JX683424 | Protein transport SEC13-like protein | 1.7 | 0 | 2 | 3.7 | Protein transport. Response to salt stress. Pentose-phosphate shunt |
| 15 | StDT33, 135 | JX683435 | Putative acid phosphatase | 3 | 2 | 2 | 7 | — |
| 16 | StDT41 | JX683414 | Class I chitinase | 2 | 2 | 2 | 6 | Chitin catabolism. Cell wall macromolecule catabolic process. Defense response to fungus. Jasmonic acid- and ethylene-dependent systemic resistance, ethylene mediated signaling pathway. Response to cadmium ion |
| 17 | StDT43 | JX845304 | Glyceraldehyde-3-phosphate dehydrogenase | 3 | 3 | 3 | 9 | Glycolysis |
| 18 | StDT45 | JX905218 | p23 Co-chaperone | 2.7 | 3 | 2 | 7.7 | — |
| 19 | StDT48 | JX683415 | Acyl-CoA-binding protein | 3 | 1 | 2 | 6 | Lipid transport. Response to cold or freezing |
| 20 | StDT49, 179 | JX951420 | Hypothetical protein | 2 | 1 | 2 | 5 | — |
| 21 | StDT50 | JX839759 | RNA recognition motif containing protein | 3 | 1 | 2 | 6 | Response to wounding. Sucrose transport. Transmembrane transport. Post-replication repair |
| 22 | StDT52 | JX683418 | LE25 protein | 1.3 | 0 | 2 | 3.3 | Embryo development ending in seed dormancy. Response to cold. Water deprivation and osmotic stress |
| 23 | StDT53, 72 | JX683419 | 60S ribosomal protein L12 | 1.7 | 2 | 2 | 5.7 | Translation. Response to cold |
| 24 | StDT55, 78 | JX951424 | Hypothetical protein | 1.3 | 1 | 2 | 4.3 | — |
| 25 | StDT56 | JX896424 | Abscisic acid and environmental stress-inducible protein TAS14 | 3 | 3 | 2 | 8 | Stress response. Response to water |
| 26 | StDT57 | JX683421 | Stress-associated protein 3 | 2 | 0 | 2 | 4 | Response to stress |
| 27 | StDT58 | JX905211 | WD-repeat protein, partial | 1.3 | 2 | 2 | 5.3 | — |
| 28 | StDT59 | JX905212 | 30S ribosomal protein S7, partial | 3 | 3 | 2 | 8 | Translation |
| 29 | StDT62, 188 | JX839758 | Abscisic stress ripening protein (ASR) partial | 1 | 1 | 2 | 4 | Response to stress |
| 30 | StDT63 | JX839746 | Aspartic protease, partial | 1 | 1 | 2 | 4 | Proteolysis. Response to salt stress. Response to cadmium ion |
| 31 | StDT64 | JX839747 | Endomembrane protein emp70, partial | 1 | 2.3 | 1.7 | 5 | — |
| 32 | StDT67 | JX683423 | 60S ribosomal protein L10-like protein | 1 | 0 | 3 | 4 | Translation |
| 33 | StDT74 | JX683425 | Fiber protein Fb11 | 1.3 | 0 | 1 | 2.3 | — |
| 34 | StDT75 | JX839748 | Short-chain alcohol dehydrogenase, partial | 0.7 | 0 | 2 | 2.7 | — |
| 35 | StDT79 | JX839749 | Dehydration responsive protein RD22, partial | 1.3 | 0 | 2 | 3.3 | Dehydration responsive |
| 36 | StDT80 | JX683426 | Ubiquitin-conjugating enzyme 2 | 1.3 | 0 | 2 | 3.3 | Photoperiodism. Flowering. Post-replication repair. Ubiquitin-dependent protein catabolism/Cellular protein modification process |
| 37 | StDT82 | JX683427 | Proteinase inhibitor I | 1 | 2 | 1 | 4 | Negative regulation of peptidase activity. Response to wounding |
| 38 | StDT85 | JX905213 | 60S ribosomal protein L27, partial | 1.3 | 0 | 2 | 3.3 | Translation. RNA methylation. Ribosome biogenesis |
| 39 | StDT87 | JX905214 | Protein transport protein Sec23, partial | 1 | 1 | 2 | 4 | Intracellular protein transport. Glucose catabolic process. Cell growth. Response to salt stress |
| 40 | StDT99, 111 | JX683433 | 40s ribosomal protein S11 | 1 | 1 | 2 | 4 | Translation |
| 41 | StDT100 | JX683430 | Ubiquitin-conjugating enzyme E2 36 | 1 | 0 | 2 | 3 | Response to iron ion and cadmium ion. Root epidermal cell differentiation. Ubiquitin-dependent protein catabolism. Post-replication repair |
| 42 | StDT102, 136 | JX839750 | Translation initiation factor eIF-1A, partial | 1.3 | 2 | 1 | 4.3 | Protein synthesis |
| 43 | StDT103 | JX845305 | Aspartic protease inhibitor 5 | 1.3 | 1 | 2 | 4.3 | Negative regulation of peptidase activity. Response to stress |
| 44 | StDT105 | JX683431 | Lipid transfer protein | 3 | 3 | 1 | 7 | Lipid transport. Nitrate transport. Response to nitrate |
| 45 | StDT110 | JX683432 | Photosystem II 10 kDa polypeptide | 2 | 2 | 2 | 6 | Photosystem II oxygen evolving complex assembly. Response to high light intensity, red light and blue light. Response to sucrose stimulus. Cysteine biosynthetic process. Regulation of proton transport |
| 46 | StDT112 | JX905215 | 25 kDa protein dehydrin | 2.7 | 1 | 2 | 5.7 | Response to stress. Response to water |
| 47 | StDT116 | JX683434 | 60S ribosomal protein L18a | 1.3 | 1 | 2 | 4.3 | Translation |
| 48 | StDT118 | JX951421 | Hypothetical protein | 1.3 | 1 | 2 | 4.3 | — |
| 49 | StDT120, 174 | JX683440 | Hypothetical protein | 1 | 1 | 2 | 4 | — |
| 50 | StDT121 | JX905216 | Glycosyltransferase gene, partial | 1.3 | 1 | 2 | 4.3 | Response to salt stress. Response to toxin. Xenobiotic catabolic process |
| 51 | StDT134 | JX839752 | UDP-glucose:glucosyltransferase | 2 | 2 | 2 | 6 | Metabolic process |
| 52 | StDT138 | JX839753 | ADP, ATP carrier protein | 1.3 | 0 | 2 | 3.3 | Transmembrane transport of purine nucleotide. Gluconeogenesis. Developmental growth. Ubiquitin-dependent protein catabolic process. Cytoskeleton organization. Photorespiration. Root hair elongation. Response to misfolded protein. Photorespiration |
| 53 | StDT141 | JX951422 | Hypothetical protein | 2.7 | 3 | 1 | 6.7 | — |
| 54 | StDT147 | JX839754 | 60S ribosomal protein L1 | 1 | 1 | 2 | 4 | Translation, RNA methylation. Cell wall modification |
| 55 | StDT149 | JX683436 | Inducible plastid-lipid associated protein | 1 | 1 | 2 | 4 | — |
| 56 | StDT151 | JX896425 | Tyramine N-feruloyltransferase 4/11, partial | 1.3 | 1 | 2 | 4.3 | Ornithine metabolic process. Response to jasmonic acid stimulus |
| 57 | StDT154 | JX845306 | ATP-dependent Clp protease proteolytic subunit | 1 | 1 | 2 | 4 | Proteolysis. Response to reactive oxygen species. Systemic acquired resistance. Regulation of protein localization. Regulation of gene expression. Regulation of defense response |
| 58 | StDT160 | JX839755 | Fructokinase 3 | 1 | 0 | 2 | 3 | Phosphorylation. d-ribose metabolic process. Cysteine biosynthetic process |
| 59 | StDT162 | JX683438 | 4F5 protein family protein | 1 | 1 | 2 | 4 | — |
| 60 | StDT171 | JX683439 | Hydrophobic protein OSR8 | 1 | 0 | 2 | 3 | Cellular response to water deprivation. Response to cold. Response to salinity. Defense response to fungus |
| 61 | StDT181 | JX683441 | Ribosomal protein L36 | 1.7 | 1 | 2 | 4.7 | Translation |
| 62 | StDT182 | JX683442 | Wound-induced protein WIN2 | 2 | 2 | 2 | 6 | Defense response to fungus, bacterium. Response to virus. Response to ethylene stimulus. Response to herbivore. Systemic acquired resistance. Response to ethylene stimulus |
| 63 | StDT184 | JX839760 | Alpha-1,4-glucan-protein synthase | 2.3 | 1 | 1 | 4.3 | Cell wall organization, Cellulose biosynthesis |
| 64 | StDT185 | JX839757 | Lipase, partial | 2 | 0 | 2 | 4 | — |
| 65 | StDT189 | JX683443 | Hypothetical protein | 2.3 | 1 | 2 | 5.3 | Transmembrane transport. Response to nematode |
| 66 | StDT190 | JX683444 | Stress enhanced protein 2 | 2 | 0 | 2 | 4 | Regulation of transcription, DNA dependent. Regulation of flower development. Photosynthesis, light harvesting. Meristem development. Vernalization. Anthocyanin accumulation in response to UV light. Plant-type cell wall modification. Post-embryonic morphogenesis. Meristem development |
| 67 | StDT200 | JX951423 | Hypothetical protein | 3 | 1 | 2 | 6 | — |
| 68 | StDT202 | JX683445 | Putative thioredoxin m2 | 2.3 | 1 | 2 | 5.3 | Cell redox homeostasis. Glycerol ether metabolic process. Response to oxidative stress. Regulation of catalytic activity |
| 69 | StDT213 | JX683446 | Hypothetical protein | 1.7 | 1 | 2 | 4.7 | — |
2.11 Quantitative RT–PCR expression analysis of the selected genes
Specific real-time PCR primers for the randomly selected genes were synthesized, and the quantitative RT–PCR expression study was performed using the cDNA synthesized from total RNA extracted from the control (untreated) and treated plants (2 h, 12 h, 24 h of drought stress). Plant preparation and drought treatment were carried out in the same manner as described above. First-strand cDNA was synthesized from 750 ng of total RNA using RocketScript Reverse Transcriptase (Bioneer, Daejeon, Korea) and oligodT (Bioneer, Daejeon, Korea) and RocketScript RT PreMix Kit (Bioneer, Daejeon, Korea). For PCR, 2 μL of cDNA were used as a DNA template with a reaction volume of 25 μL using Accupower® 2 × greenstar qpcr master mix (Bioneer, Daejeon, Korea) with the cycling conditions as follows: 95 °C for 10 min, 95 °C for 20 s and 60 °C for 45 s. The amplification reaction was carried out over 40 cycles for all the genes. RT–PCR analyses were carried out using three independent total RNA samples. Gene expression levels of each gene were normalized to actin gene (internal control) expression and were represented in graph as fold change in expression with respect to the corresponding controls (0 h of treatment).
3 Results
3.1 Optimum sorbitol concentration in drought selection media (DSM) for the selection of drought-tolerant yeast transformants
When an equal number of the control yeast cells were plated on a DSM medium containing different concentrations of sorbitol and were grown for five days, it was found that as the sorbitol concentration in the media increased, the number of colonies appearing on the plates decreased. There was no growth of the control yeast cells (Fig. 3) at a sorbitol concentration of 2.1 M and above. Hence, for functional screening, a DSM medium containing 2.1 mol of sorbitol per liter was used.
3.2 Construction of the cDNA library and transformation into yeast
The method used for the synthesis of potato cDNA, further cloning of it in the shuttle vector and the transformation of the vector into E. coli (see Materials and methods) yielded approximately 80,000 colony-forming units of E. coli transformants harboring potato cDNA inserts. We pooled all these colonies to get an unamplified library, from which plasmid was extracted and stored at –70 °C for further use in yeast transformation and functional analysis. The yeast transformation method adopted by us yielded approximately 50,000 transformants per transformation, and in the functional screening method, the product of a single yeast transformation was plated onto 10 plates containing drought selection media. Hence, approximately half a lakh yeast transformants expressing potato cDNA were screened to select the potato genes capable of imparting relatively high hyperosmotic stress tolerance to yeast.
3.3 Validation of the use of a medium containing 2.1 mol of sorbitol per liter for the functional screening of yeast cells expressing potato cDNA to select drought-tolerant yeast transformants
Based on the experiments with the control yeast cells, the concentration of sorbitol in the drought selection medium was set to 2.1 M. To further validate the use of this particular concentration for the selection of drought-tolerant yeast transformants, plated yeast (the same number, approximately 5000) cells transformed with the potato cDNA library onto DSM plates with different concentrations of sorbitol (1, 1.5, 1.8 and 2.1 M) and incubated for five days. It was found that as the concentration of sorbitol increased in the medium, the number of transformants that were grown decreased (Fig. 4). At the tested maximum sorbitol concentration (2.1 M), the number of yeast cells grown was in the range of 5 to 20 colonies per plate, which is drastically less than the number of colonies grown on media containing lower concentrations of sorbitol. Approximately 5000 colonies were plated on each plate, and 5 to 20 colonies were grown on each plate. Hence, from ten similar plates, we obtained 50 to 120 yeast transformants resistant to drought stress. The experiment was repeated three times, and we found a similar pattern. The range of 50 to 120 looks ideal in terms of ease of handling and maintaining the colonies in laboratory for further studies with tolerant yeast transformants. Hence, the use of a selection media containing 2.1 mol of sorbitol per liter (plate with DSM medium) is ideal for the selection of yeast transformants having relatively higher drought/osmotic stress tolerance, from a pool of thousands of yeast transformants expressing potato cDNA.
3.4 Yeast functional screen to select for the drought-tolerant transformants
Approximately 50,000 yeast transformants were screened for their ability to survive in a DSM medium containing 2.1 mol of sorbitol per liter. From this screening, 87 tolerant colonies were recovered. All 87 colonies were picked and streaked on the control as well as the drought selection plates, and were allowed to grow for five days. There was a clear difference in the growth of control and tolerant yeast transformants on drought selection plates, but all the colonies grew equally well on the control plates (Fig. 5). The difference in growth of control yeast and drought-resistant yeast transformants on drought selection plates can be attributed to the expression of specific potato cDNA, which might be involved in drought tolerance of potato as well as osmotic stress tolerance in yeast. The difference in the growth of control and tolerant yeast transformants on drought selection plates is quite evident from Fig. 5.
3.5 Plasmid rescue and sequence analysis
Plasmids were isolated from all 87 drought-tolerant yeast transformants, and they were back-transformed into E. coli. Plasmids were again isolated from these E. coli and specific cDNA inserts were sequenced. cDNA inserts in the plasmids of all 87 yeast colonies were identified by BLAST analysis of their sequences; their annotation and NCBI gene bank accession numbers are provided in Table 2. Fourteen different cDNA inserts were found to appear more than once (two to four times) in the total list of 87 sequences, and hence, we got 69 distinct sequences of cDNA inserts, out of which 54 are full-length and 15 are partial sequences.
3.6 Relative tolerance of the selected yeast colonies to drought, salt and high-temperature stresses
A comparison of the tolerance of the selected 69 colonies of yeast transformants expressing different cDNA sequences from potato to different stresses was carried out (Fig. 6). A relative score of tolerance is provided in Table 2. The scores ranged from 0.7 to 3. The tolerance of different yeast transformants could be compared by considering their score, where a higher score represented a greater tolerance. The difference in the tolerance between the yeast transformants indicates that the particular cDNA/gene additionally expressed in each yeast transformants improved the stress tolerance of yeast cells in different degrees. Hence, the score representing the relative tolerance of yeast cells can be taken as a reflection of the relative ability of different cDNA/genes to make yeast better able to survive stress. Thus, we can represent the relative tolerance score of each yeast transformant as the ability of that particular gene. Out of the 69 distinct genes listed in Table 2, nine genes scored the highest score of 3, three genes scored 2.7, five scored 2.3, thirteen scored 2, four scored 1.7, sixteen scored 1.3, eighteen scored 1, and one scored the lowest value of 0.7, as far as drought tolerance is concerned. With these data, we could say that the 12 genes with the highest score of 3 and 2.7 could be more useful in the drought-breeding programs of potato. The 12 genes are as follows: a glyceraldehyde-3-phosphate dehydrogenase (GPD), two dehydrins (TAS14 dehydrin and a 25 kDa protein dehydrin), a 30S ribosomal protein S7 (RPS7), a p23 co-chaperone, an acyl-CoA-binding protein (ACBP), a SUMO E2 conjugating enzyme SCE1, an acid phosphatase, a lipid transfer protein (LTP), a RNA recognition motif (RRM)-containing protein, and two genes with unknown functions. Detailed discussions on these genes are given in Section 4.1.
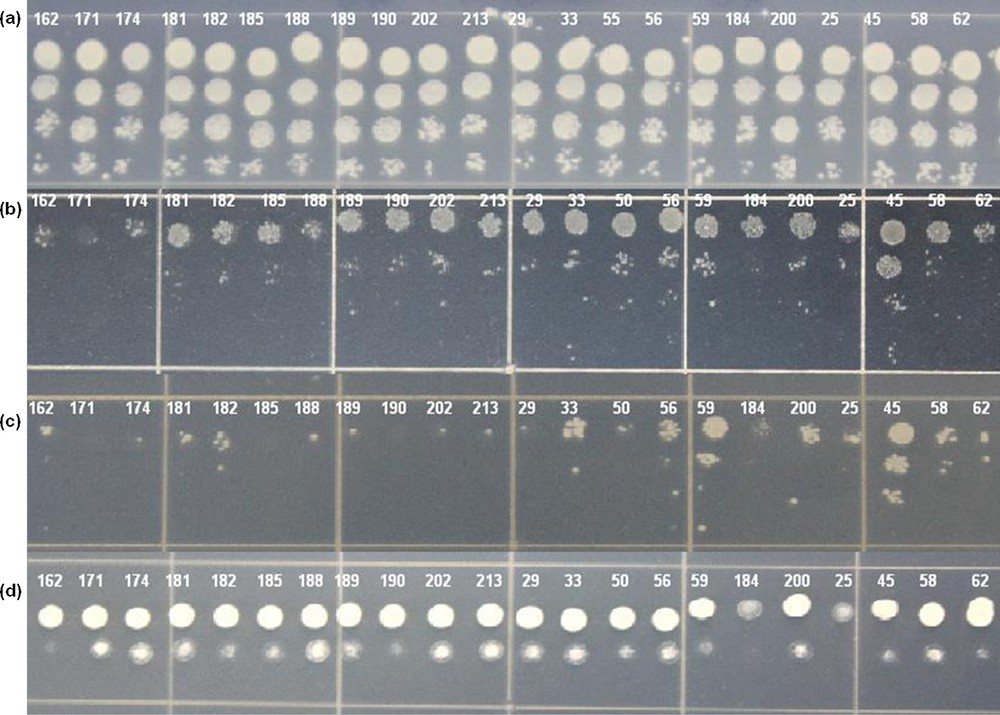
Representative figure of the relative tolerance test of the selected yeast colonies to drought (b), salt (c) and high temperature (d) stresses. Numbers in the picture represent the clone ID of the cDNA expressed by the particular yeast transformant. (a) Growth under control condition. Color online.
The scores reflecting the relative tolerance of the yeast cells expressing particular genes to salt and temperature stress are also given in Table 2. Based on these relative tolerance scores, it is possible to choose the best candidate genes, which may impart relatively better tolerance to plants in salt and high-temperature stresses. In addition, genes that might give higher tolerances under multiple stresses can also be selected based on the multiple tolerance scores listed in Table 2.
3.7 Expression pattern of selected genes in potato under drought stress
To understand the trend in the expression pattern of the identified genes in relation to drought stress, quantitative RT–PCR analysis of 17 randomly selected genes was performed. Transcript abundance of each gene in the samples collected at different time intervals of the drought stress treatment was analyzed. Gene expression levels were normalized to actin, and the values were represented in graph as fold change in expression with respect to the corresponding controls (0 h of treatment). A representative graph is shown in Fig. 7. The expression pattern of the genes had a general trend; they were found to be stress-inducible and their expression levels increased with time from 0 h to 24 h of treatment.
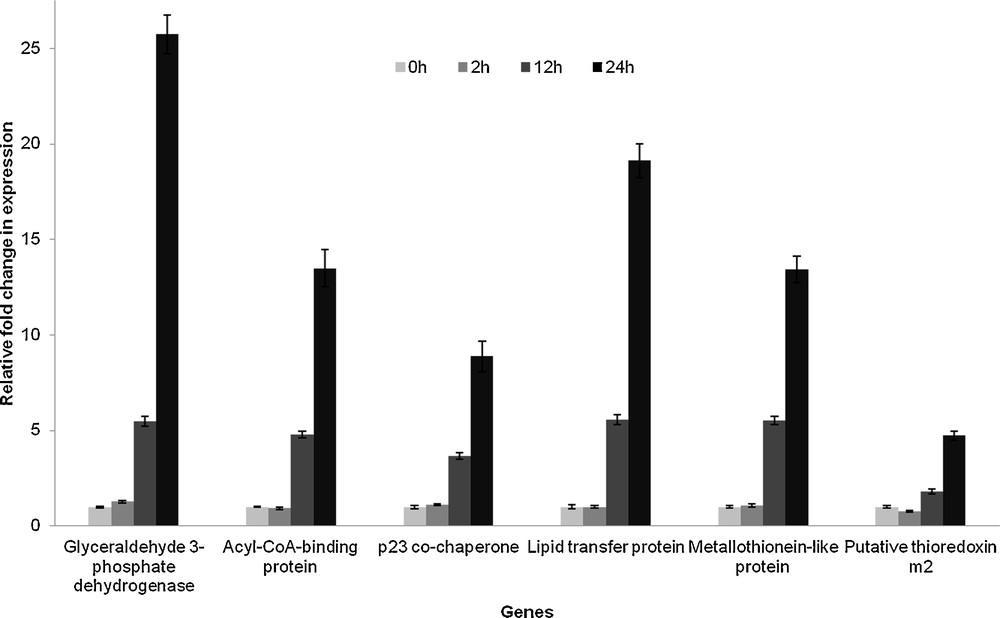
Fold change in expression level of different genes with respect to the corresponding controls (0 h treatment) in whole plant under drought stress (0 h/control/without treatment, 2 h, 12 h and 24 h, n = 3, values = ± SE).
3.8 Sequence accession numbers
Sequence data from this article can be found in the GenBank data libraries under the accession numbers: JX683406–JX683412, JX683414, JX683415, JX683417–JX683419, JX683421, JX683423–JX683428, JX683430–JX683446, JX839744–JX839750, JX839752–JX839755, JX839757–JX839760, JX845303–JX845306, JX905211–JX905216, JX905218, JX896424–JX896425 and JX951420–JX951424.
4 Discussion
Out of the 69 genes that are identified here, 61 are known to code for characterized proteins, and the remaining eight are genes with unknown functions (codes for hypothetical protein). The protein products of those genes which code for characterized protein mainly includes co-chaperone, late embryogenesis abundant proteins, environmental stress-inducible protein, carbohydrate metabolism-related proteins, RNA-binding proteins, transporters of protein, sucrose transporter, transporters of purine nucleotides, proteases, protease inhibitors, transcription factors, electron transporters of respiratory chain, enzymes involved in lipid and phospholipid metabolism, enzymes in steroid metabolism and ribosomal proteins (Table 2). The genes with unknown function could be novel determinants of stress tolerance. Biological processes of 32 out of 69 genes are in relation with various plant stress mechanisms (viz., stress response, stress tolerance, stress-related signaling etc., and those functions are bold-highlighted in Table 2). Real-time expression studies, which were carried out to understand the gene expression pattern, revealed that, in general, the expression of the genes identified in this study is stress-inducible. Expression of genes that play a role in stress response can be stress-inducible [23]; hence, the stress-inducible nature of the genes identified in this study can be taken as a sign that they have a role in the stress response of potato. However, detailed studies are needed to explore their role in drought tolerance of potato.
The primary aim of this study is to select few potato genes for the detailed investigations on their probable role in potato drought-tolerance. The criterion set to choose the genes is the relative ability of different potato genes in imparting hyperosmotic stress tolerance to yeast. Potato genes were expressed in yeast, and we used a particular scoring system to compare their ability to enhance stress tolerance of yeast; the scores of the different genes are listed in Table 2. Twelve different genes scored either of the top two scores (3 and 2.7). It is interesting to note that the different yeast cells expressing these 12 genes were among the first 17 yeast transformants showing the highest tolerance to multiple stresses (based on the multiple tolerance score, got by adding the scores of tolerance to drought, salt, and temperature, see Table 2). Hence, these 12 genes could be most useful in imparting drought stress tolerance to potato. The special characteristics of all these twelve genes are discussed below.
4.1 Important genes for further studies in potato
4.1.1 Glyceraldehyde-3-phosphate dehydrogenase (GPD)
Glyceraldehyde-3-phosphate dehydrogenase is one of the essential enzymes in glycolysis and gluconeogenesis. In potato, stress conditions and elicitor treatments are found to regulate GPD mRNA levels [24]. Jeong et al. [25] reported that yeast expressing a GPD gene of mushroom (Pleurotus sajor-caju) had a higher resistance to drought, salt, heat, and cold stresses. When the same gene was overexpressed in potato, it conferred salt stress resistance to potato cells [26]. GPD may act as a mediator of the stress-induced metabolic responses and other integrated metabolic changes during stress, and may also act to provide the additional energy needed for cellular adjustment for growth under stress, by channeling carbon away from glycerol into the pathway, leading to glycolysis and ATP formation [25]. Thus, as the GPD gene is known to have a role in stress tolerance of plants, a detailed study of this gene in potato could yield new insights into its role in stress tolerance mechanisms of the potato. The present study, using potato-GPD gene and an earlier study of the GPD gene from mushroom [25], confirmed that the yeast cells overexpressing GPD gene from plants will have greater tolerance to abiotic stresses than normal yeast cells. Hence, a yeast cell overexpressing plant GPD gene can be chosen as a positive control in future yeast-based functional screening methods for the identification of abiotic stress tolerance genes from plants.
4.1.2 Dehydrin TAS14 and 25 kDa protein dehydrin
Dehydrins or group 2 late embryogenesis abundant proteins are one of the most studied proteins that accumulate in response to water stress in higher plants [27]. Dehydrins are anticipated to play roles in stress tolerance, by protection of enzyme activity under cold and dehydration conditions [28], by acting as free-radical scavengers [29], and membrane stabilizers [30]. They also act by osmotic potential reduction, accumulation of solutes, such as sugars and K+, and their mechanism of action is connected with an earlier and greater accumulation of ABA in leaves [31]. When dehydrin TAS14 was overexpressed in tomato, its tolerance to drought and salinity stress was improved, and the effect was found to remain for a long term without affecting the growth under non-stress conditions [31]. The second dehydrin of interest, the 25-kDa protein dehydrin, is reported to be conserved in many Rhododendron species growing in temperate climates, and it plays a major role in protecting rhododendron leaves from freeze injury [32]. cDNA sequences from potato coding for both these genes are found to impart higher tolerance to yeast cells to hyperosmotic stress, giving a hint that the dehydrins from potato may also play a role in stress tolerance mechanisms.
4.1.3 30S ribosomal protein S7 (RPS7)
Being a structural component of ribosome, ribosomal proteins are generally essential for the proper growth and development of organisms. In plants, the expression of many ribosomal proteins is known to be regulated under various stress conditions [33,34]. A study by Rogalski et al. [35] showed that the chloroplast-encoded ribosomal protein L33 is necessary for plant survival under cold stress in Arabidopsis. Not many studies have been carried out to determine the exact association between stress tolerance in plants and ribosomal proteins. Hence, characterization of RPS7 in potato could provide new insights into the specific role of ribosomal proteins in stress tolerance of plants.
4.1.4 p23 Co-chaperone
Chaperones are important protein molecules for homeostasis in cells under both optimal and adverse growth conditions [36]. Co-chaperons are proteins that function in association with chaperon molecules. Heat-shock protein 90 (HSP90), is a well-known chaperone that assists polypeptides in folding, and prevents their unproductive interactions [37]. p23 is one of the three co-chaperones of HSP90. The function of HSP90 and many co-chaperones is conserved among fungi, animals, and plants [38,39]. As p23 co-chaperones act in association with HSP90, its role in stress tolerance of plants is assumed to be important. In this study, yeast cells expressing potato p23 co-chaperone gene were found to have an improved tolerance to hyperosmotic stress. Considering all these facts, we expect that the gene coding for p23 co-chaperone can play a significant role in potato drought tolerance.
4.1.5 Acyl-CoA-binding protein (ACBP)
Acyl-CoA-binding proteins play a role in phospholipid metabolism, plant developmental processes, plant stress responses (heavy metal resistance, oxidative stress, freezing tolerance and pathogen resistance), vesicular trafficking, membrane biogenesis, and signaling pathways [40,41]. ACBPs are known to interact with an ethylene-binding protein (AtEBP) [42,43], which is a plant transcription factor involved in biotic and abiotic stresses, including pathogen defense, hormone signaling, and tolerance to drought, cold and cadmium [44]. Thus, ACBPs may play biological roles related to plant defense and ethylene signaling through the interaction with AtEBP [42,43]. ACBPs are induced by biotic or abiotic stresses, such as those caused by heavy metals, freezing, oxidative stresses and pathogen infections [43,45–47]. Altered expression of these ACBPs is found to change the tolerance of a plant to the corresponding stresses, indicating that they are directly or indirectly involved in protecting the plants from various adversities [41]. Considering all these information from earlier studies, together with the indications from our yeast-based screening study, ACBPs from potato can be expected to play a significant role in stress tolerance of potato.
4.1.6 SUMO E2 conjugating enzyme SCE1
In cells, the SUMO E2 conjugating enzyme SCE1 is involved in the SUMOylation process [48]. In rice seedlings, expression levels of a putative gene encoding SUMO E2 conjugating enzyme SCE1 was found to change with temperature stress [49]. Using yeast two-hybrid analysis, Nigam et al. [49] showed that SCE1 protein physically interacts with another important stress-related protein, OsFKBP20 (FK506-binding proteins), and proposed that SCE1 and OsFKBP20 proteins act in concert to mediate stress response in rice plants. SUMOylation status of the proteins involved in stress is known to alter as they travel between the nucleus and the cytoplasm [50]. The SUMO conjugation pathway is implicated in the activation of a chain of nuclear regulatory proteins, the activities of which may be involved in plant stress signaling [49]. Considering all these facts together with the findings from this yeast-based study, SUMO E2 conjugating enzyme SCE1 from potato may also play a significant role in stress tolerance.
4.1.7 Acid phosphatase
The activity of acid phosphatase is known to increase in a few plants, such as Alfalfa [51] and Pisum sativum [52], upon salt and drought stress treatment. Acid phosphatase act to maintain a certain level of inorganic phosphate in plant cells under conditions of stress [53,54]. According to Ehsanpour and Amini (unpublished data), when a plant increases its acid phosphatase activities under stress conditions, it becomes more resistant to stress. However, not much is known about its role in stress tolerance. In our study, yeast cells overexpressing an acid phosphatase gene from potato are found to have improved tolerance to hyperosmotic stress. A detailed study on potato acid phosphatases may bring new insights into its role in the stress tolerance of plants.
4.1.8 Lipid transfer protein (LTP)
Lipid transfer proteins are ubiquitous lipid-binding proteins in plants, which are involved in various stress responses [55]. Expression of LTPs was found to be differentially regulated by various stresses, such as high-temperature [56], cold [57], salt [58], and heavy metal stresses [59]. LTPs from various plants are known to have antibiotic properties [60–63]. Arabidopsis and tobacco plants overexpressing the LTP gene from Hordeum vulgare were found to have enhanced tolerance to bacterial pathogens [64]. Arabidopsis plants overexpressing LTP of Capsicum annum are reported to have increased salt and drought tolerance [65], indicating that LTP plays important roles in osmotic stress tolerance in plants. Considering all these findings, together with the better performance of LTP gene from potato in making yeast cells tolerant to drought, LTP is a possible candidate gene for plant multiple stress tolerance breeding.
4.1.9 RNA recognition motif (RRM)-containing protein
RRM-containing proteins play important roles in the processing of RNA and regulation of protein synthesis [66]. Arabidopsis RRM-containing RNA-binding proteins are involved in the adaptation of plants to various environmental conditions [67]. RRM is the principal RNA-binding domain of such protein, and is the most variable region of the RRM domain, probably containing determinants of target specificity [68]. Thus, the characterization of the RRM sequence of the selected RRM-containing protein may help us to predict its targets and functions. A detailed characterization of this gene in plants could bring more insight into its role in stress tolerance.
4.1.10 Two genes with unknown functions
Two cDNA sequences from potato, which have open-reading frames of 888 bp (GenBank accession number JX951422) and 411 bp (GenBank accession number JX951423) have been found to impart relatively high drought-stress tolerance to yeast cells. The potato genome sequencing consortium classified these sequences as conserved genes of unknown function. Thus, a study to characterize their roles in stress tolerance of potato would be a novel and valuable undertaking, and could provide new insights into the stress tolerance mechanism(s) of plants.
4.2 Most important candidate genes for the improvement of multiple abiotic stress tolerance of potato
The multiple abiotic stress tolerance score of each gene was calculated by adding up the relative tolerance score of the particular gene to drought, salt and high temperature stresses. Scores of multiple abiotic stress tolerance of genes are listed in Table 2. Based on the scores, it is possible to say that six genes, which scored the first four highest values (9, 8, 7 and 6.7) of multiple stress tolerance, could be more useful in breeding programs of potato for enhancing multiple abiotic stress tolerance. These six genes code for glyceraldehyde-3-phosphate dehydrogenase, abscisic acid and environmental stress-inducible protein TAS14, 30S ribosomal protein, putative acid phosphatase, lipid transfer protein and a hypothetical protein. It is interesting to note that all these six genes appeared in the selected list of most important candidate genes for drought-breeding program too.
5 Conclusion
The aim of this study was to select a few potential drought-tolerance genes for a detailed investigation of their possible involvement in potato drought tolerance. The criterion by which we chose to select among the different potato genes is their relative ability to impart hyperosmotic stress tolerance to yeast. Sixty-nine potato genes are identified as capable of enhancing hyperosmotic stress tolerance in yeast, from which 12 have been identified as being likely of great importance. A few genes reported here had previously been known as playing roles in osmotic stress tolerance in yeast as well as in drought tolerance in plants, ensuring the robustness of the method for identifying potential drought-tolerance genes that are common in plants.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgement
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008182), Rural Development Administration, Republic of Korea.


