1 Introduction
Tomatoes are one of the most popular and widely grown vegetables in the world and can be grown either in the field or under greenhouse conditions.
Due to increasing demand, tomato has a great potential for increased commercialization. Damping-off is one of the worst diseases of tomato occurring in the nursery and can kill both germinating seeds and young seedlings. Several fungi that are widely distributed in soils can cause this disease, including Rhizoctonia solani, Pythium spp., Phytophthora spp., Sclerotinia spp. and Fusarium spp. Strategies to control soil-borne diseases are limited because cultivars with complete resistance are not available [1]. The control of the soil-borne pathogens is difficult because of their ecological behavior, their extremely broad host range and the high survival rate of resistant forms, such as chlamydospores and sclerotia under different environmental conditions. Many research studies have shown that biological control offers an environmentally friendly alternative to protect plants from soil-borne pathogens [2–4]. Although the number of biocontrol products is increasing, these products still represent only a very small proportion of fungicides [5]. In recent years, several bacterial and fungal antagonists against soil-borne plant pathogenic fungi have been described [6,7].
Application of biocontrol agents (BCAs) or plant growth-promoting rhizobacteria (PGPR) is considered as an important approach in crop protection against plant pathogens. Several microbes have been studied extensively as BCAs against various phytopathogens and these also showed plant growth promotion activity [8,9]. In particular, the application of fluorescent Pseudomonads has received particular attention because of their catabolic versatility, excellent root-colonizing abilities, and production of a wide range of antifungal metabolites. Several authors [10–13] reported that Pseudomonas sp. isolates represent potent BCAs against P. ultimum, the causal agent of corn and tomato damping-off, respectively.
The objectives of this study were to select and characterize fluorescent Pseudomonas strains with antagonistic activities against phytopathogenic fungi of vegetables in vitro, and to evaluate the effect of one strain, Psf5, on tomato growth as well as its potential for controlling tomato damping-off caused by S. sclerotiorum.
2 Materials and methods
2.1 Isolation of fluorescent Pseudomonas strains
Fluorescent Pseudomonas spp. were isolated from the rhizosphere of healthy tomato (L. esculentum Mill.) and pepper (Capsicum annuum L.) plants from three regions of Gabès, Tunisia. Non-rhizosphere soil was removed from the root system of the plants.
Roots were excised and placed into 10 mL of a sterile 0.9% NaCl solution and vortexed for 10 min in order to detach the associated rhizosphere soil. Serial dilutions of the resulting root wash were plated on King B medium (KB) [14] supplemented with ampicillin (100 mg/mL) and cycloheximide (75 mg/mL) [15]. Plates were incubated at 28 °C for 24–48 h; at which time the fluorescent colonies were observed under UV light (354 nm). To obtain the most abundant bacteria from each sample, selection of strains showing fluorescence and different colony morphology was performed from the highest dilutions. All bacterial cultures were stored at –20 °C in tryptic soy broth (TSB) supplemented with 20% (v:v) glycerol.
2.2 Identification of antagonistic bacteria
Bacterial characterization was carried out on the basis of colony morphology, Gram stain, oxidase test, production of acids from 1% glucose in oxidation/fermentation (OF) basal medium [16], and analysis with the API 20NE biochemical test. Molecular identification of antagonistic strains that were used in this study was made by sequencing the 16S rRNA gene (rrs). Amplification was carried out by PCR with primers F667-pA-rrs AGA GTT TGA TCC TGG CTC AG and F668-pH-rrs AAG GAG GTG ATC CAG CCG CA designed by Bruce et al. [17]. Standard PCR conditions were 1 min DNA denaturation at 94 °C, 1 min annealing at 57 °C, and 1 min extension at 72 °C for 35 cycles. The 16S rDNA sequences were compared to sequences in the GenBank database with the Basic Alignment Search Tool (BLAST) [18].
2.3 Phytopathogenic fungi
Fungal phytopathogens (Sclerotinia sclerotiorum, Fusarium solani and Alternaria alternata) were originally isolated from tomato plants exhibiting symptoms of tomato damping-off. Infected roots were washed with running tap water to remove any soil remains, and then cut into small pieces before being dipped in sodium hypochlorite solution (2%) for two minutes for surface sterilization. These plants were then washed several times with sterile distilled water, dried between sterilized filter paper, then placed on PDA medium with and without antibiotics. The plates were incubated at 28 °C and scanned daily for fungal development. Preliminary microscopic examination of the fungi isolated showed that they could be classified under three genera: Fusarium, Alternaria, and Sclerotinia. Fusarium and Alternaria spp. isolates were purified by plating single conidial spores [19], while Sclerotinia was purified using the hyphal tip technique [20]. Representative isolates were maintained on PDA slants for further studies. Isolated fungi were identified according to their morphological features as described by Booth et al. [21], Dhingra and Sinclair [22], and Barnett and Hunter [23].
2.4 Molecular identification of fungi
DNA was isolated from fungal mycelia (0.02 g) by grinding with quartz sand, and extraction following the Qiagen DNeasy Plant kit protocol. The ITS region of the fungal DNA was amplified using the fungal specific primer set: ITS1-F (CTT GGT CAT TTA GAG GAA GTA A) and ITS4 R (TCC TCCGCT TAT TGA TAT GC) as described by White et al. [24]. Amplifications were performed in 100 μL PCR reaction tubes containing 5 μL of DNA, 10 μL of 10× thermophilic buffer, 8 μL of MgCl2 (25 mM), 4 μL of dNTP (10 mM), 10 μL of ITS1-F (10 μM), 10 μL of ITS4 (10 μM), 52 μL of ddH2O and 1 μL of Taq (Promega, 10 u/μL). Amplifications occurred in an Eppendorf Mastercycler Thermal Cycler with the following program for 40 cycles: initial denaturation temperature 94 °C for 90 s, melt temperature 95 °C for 35 s, annealing temperature 55 °C for 55 s, extending temperature of 72 °C for 1 min, final extension temperature 72 °C for 10 min, hold temperature 4 °C [25]. The ITS bands were identified by gel electrophoresis on 2% agarose. The molecular weights of each ITS fragment were determined using GelPro Express software and a 100 bp ladder. For DNA sequencing, excess primers and buffers were removed from the amplified ITS fragment according to the procedures given in the Mo Bio PCR DNA purification kit. DNA concentration, as determined by fluorescence, was done according to procedures in the Sigma DNA Quantification Kit and ranged between 2 ng/μL and 100 ng/μL. The fungal ITS fragment was prepared for sequencing according to the Beckman CEQ DTCS kit. The target DNA concentration was 45 ng/50 μmol. The forward and reverse sequences for each fungal species were aligned with Clustal W [26] validated visually and a consensus sequence was generated also with Clustal W. The identity of the consensus sequence was confirmed using BLAST search relative to known sequences in NCBI Genbank.
2.5 Evaluation of strains for in vitro biological control
2.5.1 Antagonism in dual culture
Fluorescent Pseudomonas was tested against S. sclerotiorum., A. alternata and F. solani in plate bioassays. A. alternata and F. solani were cultivated in PDA at 28 °C. Conidia were harvested from the surface of plates by flooding the 10-day-old cultures with 9 mL of sterilized distilled water and gently scraping with a sterilized glass rod; conidial concentration was determined with a Neubauer chamber [27]. Plates containing the media to be tested (KB, PDA) were prepared. Then, an agar over-layer containing the target fungus, immobilized at a concentration of 104–105 conidia/mL, was placed on the medium. The methodology described by Montesinos et al. [28] was followed in order to prepare the overlay, using 0.7% agar. Four milliliters of the medium were placed in screw-capped test tubes, which, once sterilized, were kept inside a water bath at 40 °C. Next, 10 mL of a target conidia suspension were added to each test tube, which were vortexed; the content of each tube was then homogeneously distributed on a plate containing the same culture medium. The bacterial strains tested were sown by gently touching the agar surface with a sterile toothpick, previously inoculated by touching the surface of a single colony. Plates were incubated for 72 h at 28 °C. The degree of inhibition in each medium was determined by measuring the halo around the bacterial strain without fungal growth. Six replicates were considered for the value of the inhibition halo. For screening for potential antagonism against S. sclerotiorum, mature sclerotia were removed from the surface of 15-day-old cultures with a sterile forceps, and four samples were immediately placed around the edges and one in the center of a plate 24 h after the stab-inoculation of four bacterial strains. The experiment was conducted twice.
2.5.2 Mycelial growth inhibition
The bacterial strains were streaked on 1/3 of a Petri plate containing 25% TSA, KB or PDA. A mycelial disc (9 mm diameter) of a 8–15-day-old culture of an actively growing target fungus was equidistantly placed on the opposite side of the Petri plate 48 h after inoculation of the strain. Plates were incubated for seven days at 28 °C. The plates with fungal pathogens on one side that were not inoculated with bacterial strains served as controls. For each fungal colony, two diameters, measured at right angles to one another, were averaged to find the mean diameter for that colony. The mean diameter of fungal growth in the presence of each strain was compared to that of the control cultures in order to determine the inhibition percentage. All fungal colony diameters were determined by using three replicates for each strain on each medium. Plates were incubated for seven days at 28 °C. The fungal colony diameter was determined by using three replicates for each strain on each medium. The plates with tested fungi on one side that were not inoculated with bacterial strains served as controls. The experiments were conducted twice.
2.5.3 Production of hydrolytic enzymes
Proteolytic activity was detected by inoculating the strains on a medium composed of 1% casein and 2.3% agar dissolved in Castañeda medium [29]. Plates were incubated for 48 h at 28 °C. Casein hydrolysis was detected by the formation of a whitish (opaque halo coagulated casein) around a translucent area (totally hydrolyzed casein), surrounding the colony. To determine the cellulolytic activity, carboxymethyl cellulose (CMC) was incorporated at 0.1% into the YEMA-0.2% mannitol agar plates. Colonies were grown for three days at 28 °C and washed off with water. The plates were then flooded with 0.1% (wt:vol) Congo Red in water for 15 min, washed for 10 min with 1 M NaCl, and then washed for 5 min with 5% acetic acid. Degradation of CMC was observed as clearings (reduction of staining) [30]. All the hydrolytic tests were performed twice.
2.5.4 Assay of chitinases
P. fluorescens strains were cultured at 28 °C for 96 h on a rotary shaker in 250 mL conical flasks containing 50 mL of chitin–peptone medium (glucose 0.5%, peptone 0.2%, colloidal chitin 0.2%, K2HPO4 0.1%, MgSO4·7H2O 0.05%, and NaCl 0.05%, pH 6.8). The cultures were centrifuged at 12,000 g for 20 min at 4 °C and the supernatant was used as enzyme source. Colloidal chitin was prepared from crab shell chitin according to Berger and Reynolds [31]. The reaction mixture contained 0.25 mL of enzyme solution, 0.3 mL of 1 M sodium acetate buffer (pH 5.3) and 0.5 mL of colloidal chitin (0.1%). The reaction mixture was incubated at 50 °C for 4 h in a water bath. Chitinase activity was determined by measuring the release of reducing sugars by the method of Nelson [32]. One unit of chitinase was determined as 1 nmol of GlcNAc released per minute per mg of protein. Protein content in all the samples was determined as described by Bradford using bovine serum albumin as the standard.
2.5.5 Production of siderophore by P. fluorescens
The chrome azurol S (CAS) method described by Alexander and Zuberer was used for screening strains for siderophore production. Plates were incubated at 28 °C for five days, and microorganisms exhibiting an orange halo were considered to be producers of siderophores.
2.5.6 Identification and quantification of indole-3-acetic acid (IAA) in culture supernatant of strain Psf5
Strain Psf5 was grown in nutrient broth (NB). Then, 20 mL were taken during the late exponential growth phase (24 h) for identification and quantification of IAA, which were carried out by high performance liquid chromatography–mass spectrometry (HPLC–MS). A 100-ng 2H5-IAA deuterated internal standard was included.
2.6 Evaluation of selected strain Psf5 for growth promotion and biological control
2.6.1 Preparation of fungal and bacterial inocula and treatment of seeds
Cultures of S. sclerotiorum were maintained on PDA, on which brown sclerotia were formed within 8–10 days. Pathogen inocula added to sterile mixture consisted of 30-day-old sclerotia, which were dislodged from the surface of plates and used immediately [33]. Plastic pots (15 cm diameter; 25 cm height) were filled with 600 g of sterile mixture (soil:sand:perlite at 2:1:1 w/w/w), previously sterilized by heating at 180 °C for 2 h on four consecutive days. Each pot was then moistened with sterile distilled water and infested in the mixture surface with 30 mg of sclerotia. Pots were kept for 10 days in a growth chamber under controlled conditions: 16 h light at 28 ± 2 °C, 8 h dark at 16 ± 2 °C (light intensity of 220 μE m2/s). After incubation, tomato seeds were surface-disinfected for 10 min in 5% sodium hypochlorite solution (60 g/L of active chlorine), washed ten times in sterilized distilled water, and air dried [34]. Then, 10 g of seeds were soaked for 30 min in 2.5 mL of a 109 CFU/mL aqueous cell suspension of strain Psf5. The bacterium was prepared by growing by shaking (80 rpm) in KB broth for 48 h at 28 °C. Then, eight inoculated seeds were placed into the mixture surface in each pot. The four treatments were:
- • non-infested, non-bacterized healthy control (treated with sterile distilled water);
- • non-infested, bacterized with Psf5 alone;
- • infested with S. sclerotiorum, non-bacterized control;
- • infested with S. sclerotiorum, bacterized with Psf5.
The pots were incubated in a growth chamber under the conditions described above. Damping-off was determined by counting the total healthy stand after 40 days, compared to the non-infested control plants. Shoot and root dry weights (72 h at 70 °C) were recorded from twenty randomly selected plants from each treatment. The pots were arranged in a completely randomized design. The experiment was performed twice, each with six replicates per treatment.
2.6.2 Tomato rhizosphere colonization
Survival of strain Psf5 in the rhizosphere of tomato plants from treatments 1 and 4 was determined according to a modification of the procedure described by Landa et al. [35], 10, 25, and 40 days after sowing. For determining colony count at 1 h (zero time), 1 g of rhizosphere mixture was collected from the surroundings of a seed and placed into 9 mL of a sterile 0.9% NaCl solution. Serial dilutions of the suspension were vortexed and plated onto a 25% TSA medium. Plates were incubated for 48 h at 28 °C; the developed colonies were counted and the number of CFU/g of the mixture was calculated.
Also, a seedling from each treatment was carefully removed from a pot at 10, 20, 30, and 40 days and roots were gently shaken to remove all but the tightly adhering potting mixture. One gram of the adhering rhizosphere mixture was collected and placed into 9 mL of a sterile 0.9% NaCl solution. Serial dilutions of the suspension were vortexed and plated onto 25% TSA medium. Plates were incubated for 48 h at 28 °C. The developed colonies from each treatment were counted and the number of CFU/g of mixture was calculated.
2.7 Statistical analyses
The data were analyzed by using analysis of variance (ANOVA). When ANOVA showed treatment effect, the Least Significant Difference (LSD) test was applied to make comparisons between the means at P < 0.05. All data were subjected to statistical analysis using Statgraphics plus software for Windows V4.1 (Statistical Graphics Corp., Maryland, USA).
3 Results
3.1 Isolation of microorganisms and screening potential antagonists
Twelve bacterial strains were obtained from tomato roots. All of the strains were Gram-negative rods, oxidase-positive and capable of metabolizing glucose in an oxidative form. Sequencing of 16S rDNA and comparison with GenBank database of sequences revealed that antagonistic strains belong to the species Pseudomonas fluorescens (five strains), P. putida (five strains), and P. aeruginosa (two strains). Recovered bacterial strains were tested for their antagonistic ability against the phytopathogenic fungi A. alternata, S. sclerotiorum, and F. solani. As a result, the bacterial antagonistic effect in the dual culture assay depended both on the target pathogen and the culture media used; moreover, the influence of the composition of the medium was observed against all fungi. The strongest in vitro antagonism against A. alternata was observed on 25% TSA, while the higher inhibitory activity against F. solani and S. sclerotiorum was observed on KB.
3.2 Mycelial growth inhibition
The observed in vitro inhibition of mycelial growth also varied with the culture medium and the target pathogen. The inhibitory effect on the mycelial growth of A. alternata was higher on 25% TSA. All the tested strains resulted in > 70% inhibition on 25% TSA, > 47% on KB and ≤ 20% on PDA. As observed for the germination inhibition assay, mycelial growth inhibition of F. solani and S. sclerotiorum by all the recovered strains was more effective on KB. However, the growth of the fungal pathogens was barely inhibited in the presence of the strains on PDA (Table 1).
Effect of three different media on the mycelial growth of S. sclerotiorum (S. s), A. alternata (A. a) and F. solani (F. s), measured as diameters of the fungal colonies in the presence of each Pseudomonas bacterial strain, compared to the control plate.
| Bacterial strain | Inhibition percentage of mycelium growth | ||||||||
| PDA | TSA | KB | |||||||
| S. s | A. a | F. s | S. s | A. a | F. s | S. s | A. a | F. s | |
| P. fluorescens Psf1 | 4.95bc | 14.57bc | 5.40d | 1.99c | 72.84ab | 4.36c | 11.47cd | 50.58ns | 12.06cdef |
| P. fluorescens Psf2 | 19.90a | 13.54bc | 20.27a | 51.19a | 79.00ab | 28.38a | 59.00a | 57.71ns | 43.21a |
| P. fluorescens Psf 3 | 5.37bc | 11.98bc | 8.10c | 1.75c | 82.11a | 4.79c | 20.07bc | 54.50ns | 9.54ef |
| P. fluorescens Psf 4 | 5.78bc | 13.54bc | 5.40d | 17.19b | 77.48ab | 3.05cde | 8.19cd | 55.82ns | 11.05cdef |
| P. fluorescens Psf 5 | 7.02bc | 15.62bc | 7.66c | 2.39c | 71.80ab | 2.17ef | 25.81bc | 50.58ns | 9.04f |
| P. putida Pp1 | 7.43bc | 20.31b | 3.60d | 12.39b | 60.65b | 2.62de | 9.01cd | 41.43ns | 12.55bcde |
| P. putida Pp 2 | 2.88cd | 9.89c | 8.10c | 4.39c | 73.86ab | 4.79c | 2.45d | 47.98ns | 10.55def |
| P. putida Pp 3 | 2.47cd | 15.10bc | 9.01c | 1.79c | 75.40ab | 1.74f | 3.68d | 44.05ns | 14.06bc |
| P. putida Pp 4 | 6.61bc | 7.81c | 5.40d | 1.59c | 80.04ab | 1.74f | 1.63d | 41.43ns | 9.54ef |
| P. putida Pp 5 | 6.65bc | 7.86c | 5.80d | 2.39c | 72.80ab | 2.17ef | 25.81bc | 52.58ns | 9.04f |
| P. aeruginosa Pa1 | 2.83cd | 9.82c | 8.10c | 4.32c | 73.80ab | 4.73c | 2.43d | 47.94ns | 10.50def |
| P. aeruginosa Pa2 | 7.43bc | 16.14bc | 7.66c | 1.00c | 75.93ab | 3.48cde | 24.58bc | 53.19ns | 13.56bcd |
3.2.1 Production of hydrolytic enzymes
Three strains (Psf 4, Pp1, and Pa2) showed protease activity, whereas none of them produced cellulase or chitinase. Additionally, all of the strains were able to respond to iron limitation, producing siderophores in CAS medium. Out of the twelve strains isolated from the root system of tomato plants, strain P. fluorescens Psf5 was selected for further study based on its in vitro inhibitory activity in the antagonism in dual culture as well as in the mycelial growth inhibition assays against phytopathogenic fungi of tomato, in particular against S. sclerotiorum.
3.2.2 In vivo biological control of S. sclerotiorum
The antagonistic strain Psf5 was evaluated for growth promotion of tomato plants and biological control of S. sclerotiorum in in vivo tests. In S. sclerotiorum infested pots, the inoculation of the seeds with Psf5 strain improved the seedling stand by 38% and increased both shoot and root dry weight over the control plants by 120 and 110 mg, respectively (Fig. 1). No evident differences between bacterized seeds and control seeds were observed in non-infested potting mixture when recording plant stand; however, inoculation of seeds with Psf5 increased (P < 0.05) root dry weight by 80 mg. The increase in root dry weight may be due to phytohormone-like substances. To verify the presence of IAA, the supernatant of the strain Psf5 was subjected to further HPLC analysis. In the chromatogram of the supernatant of the strain Psf5, a large peak was detected with the same retention time as that of authentic IAA (Fig. 2) The strain Psf5 produces indole-3-acetic acid (IAA) at 5.10 mg/mL (without addition of tryptophan to culture medium) after 24 h of incubation. Although inoculation with strain PF5 increased shoot dry weight by 30 mg, when compared to healthy controls, differences were not significant (Fig. 1). fluorescent Pseudomonas morphologically similar to Psf5 reached a population density of 107–108 and 106–107 CFU/g mixture after ten and forty days of experimentation, respectively, in the bacterized with Psf5 alone treatment (Fig. 3). Colony counts performed from non-infested, non-bacterized control plants revealed an absence of colonies morphologically similar to Psf5.
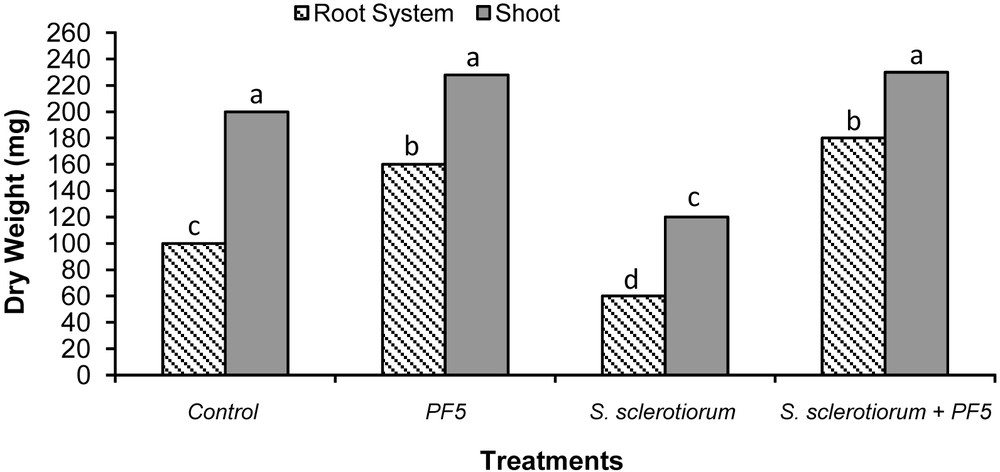
Biocontrol activity of the Pseudomonas fluorescens strain Psf5 against S. sclerotiorum on tomato plants. Tomato root and shoot dry weights were measured 40 days after the beginning of the test. The data represent the average of two different experiments. Bars for each plant fraction with different letters are significantly different according to the LSD test (P < 0.05).
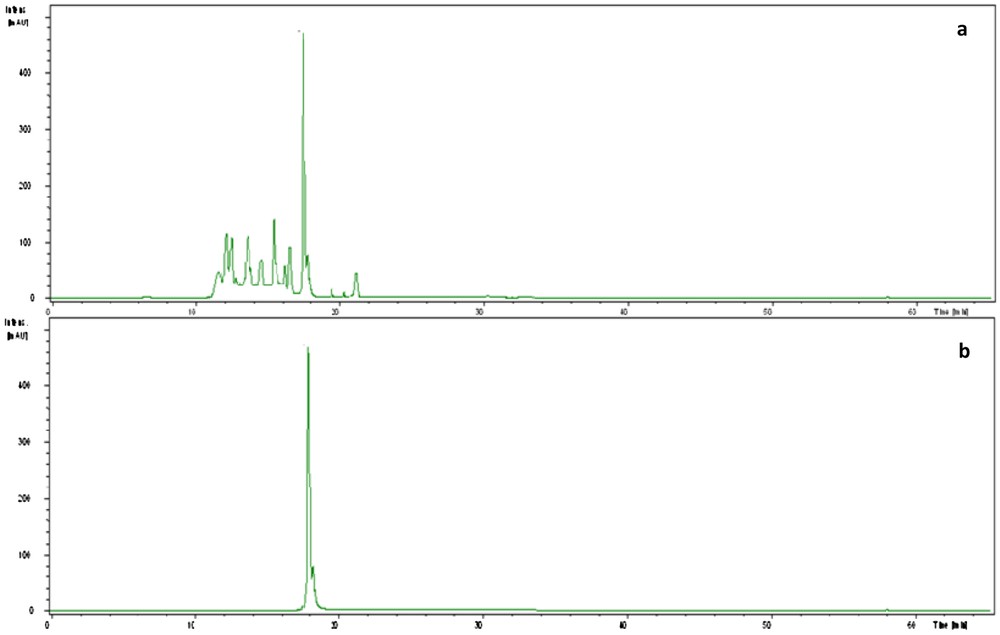
HPLC chromatogram of the supernatant of Pseudomonas fluorescens strain Psf5 (a) and the authentic IAA (b).
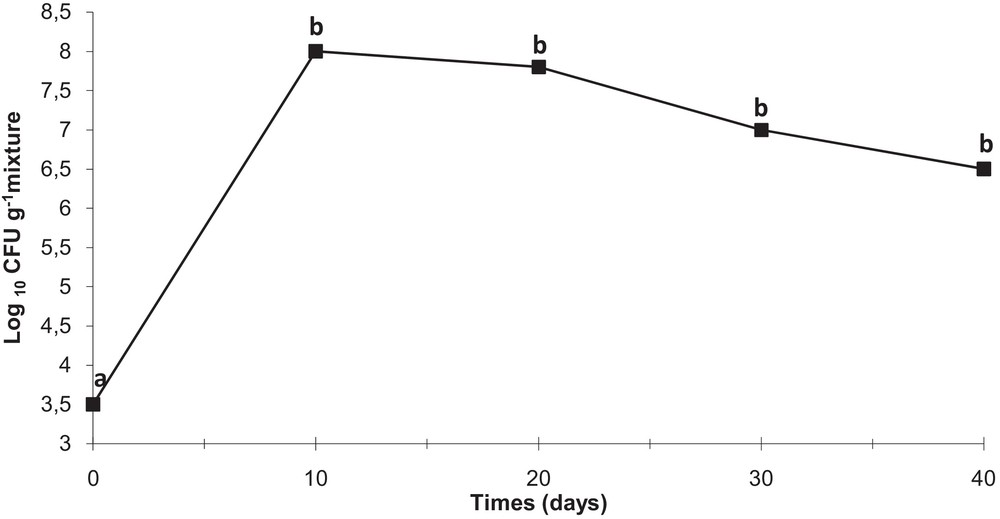
Persistence of strain Psf5 in the rhizosphere of tomato plants. For determining colony count at 1 h (zero time), 1 g of rhizosphere mixture was collected from the surroundings of a seed and placed into 9 mL of a sterile 0.9% NaCl solution. Serial dilutions of 1 g of rhizosphere mixture, collected from the rhizosphere surroundings on seeds or roots of one plantlet were plated and incubated for 48 h; the developed colonies were counted and the number of CFU per gram of mixture was calculated. Different small letters at a given time indicate significant differences between treatments at P < 0.05.
4 Discussion
The use of biological control based on natural microorganisms offers a powerful alternative to synthetic chemical control of plant diseases. In fact, the abuse of chemical control agents, such as pesticides or fungicides, to cure or prevent plant diseases has often been reported to bring about a wide array of pernicious effects, particularly on plant, soil, environment, and, ultimately, human beings. Soil-borne fluorescent pseudomonads have been widely used for the control of fungal disease as they can enhance plant growth and yield apart from suppressing the growth of the pathogen. In view of this continuing search for new natural alternatives to chemical control agents, the present study was undertaken to investigate the potential use of Pseudomonas spp. strains for the biological control of phytopathogenic fungi of vegetables in vitro, as well as its potential for controlling tomato damping-off caused by S. sclerotiorum.
The twelve Pseudomonas strains evaluated in this study were isolated from rhizosphere of healthy tomato plants from three regions of Gabes, Tunisia. Williams and Asher [36] concluded that the methods employed to isolate rhizobacteria play an important role in identification of potential biocontrol agents, and that the strains should be from the rhizosphere of the target crop.
Rhizospheric Pseudomonas strains were reported to be effective for the control of a wide range of fungal and bacterial diseases [37]. Bacteria of the genus Pseudomonas comprise a large group of the active biocontrol strains as a result of their general ability to produce a diverse array of potent antifungal metabolites. These include simple metabolites, such as 2,4-diacetylphloroglucinol, phenazine-1-carboxylic acid and pyrrolnitrin [3-chloro-4-(2′-nitro-3′-chlorophenyl)-pyrrole], as well as the complex macrocyclic lactone, 2,3-de-epoxy-2,3-didehydra-rhizoxin. Pyrrolnitrin is active against Rhizoctonia spp., Fusarium spp., and other plant pathogenic fungi, and it has been used as a lead structure in the development of a new phenylpyrrole agricultural fungicide [38]. Several antibiotic-like substances have been identified, including bacteriocins and phenazine antibiotics [39].
Molecular identification revealed that antagonistic strains belong to the species Pseudomonas fluorescens (five strains), P. putida (five strains) and P. aeruginosa (two strains). Antagonistic properties of strains tested in vitro were influenced by culture medium composition, the fungal pathogen, and its growth stages. Our results are in agreement with those of Borowicz and Saad Omer [40], who proposed that differences between media could result in alterations of the metabolites produced, or of their relative concentrations. Moreover, the type of medium used to grow both bacteria and fungi in studies of biological control affects the interaction of the organisms [41] Preliminary findings from biological control experiments carried out in vitro indicated the ability of the majority of antagonists to inhibit the phytopathogenic fungi tested. Moreover, P. fluorescens strains were found to be the most efficient by exhibiting the highest inhibition. Strains of P. fluorescens showed known biological control activity against certain soil-borne phytopathogenic fungi. In fact, Rajappan and Ramaraj [42] evaluated the in vitro efficacy of P. fluorescens against the cauliflower wilt pathogen F. moniliforme. Furthermore, Pseudomonas strains controlled stalk rot of maize associated with P. aphanidermatum and F. graminearum at the seedling stage [43]. Also, Janisiewicz and Roitman [44] have also reported that blue mold and grey mold of apples and pears could be controlled by Pseudomonas. Fluorescent Pseudomonas strains were also found to be effective against Sclerotium rolfsii [45] under greenhouse conditions in limiting groundnut and collar rot incidence.
Enzymatic degradation of the cell wall of fungal pathogens by biocontrol agents has been reported [46]. In this work, protease, cellulase, and chitinase production were assayed. Protease production was the only exoenzymatic activity detected in the Pseudonmonas spp. strains tested. Based on a significant in vitro antagonistic effect against phytopathogenic fungi A. alternata and S. sclerotiorum, a potential biocontrol agent, strain Psf5, was selected for a future evaluation of its ability to suppress fungal pathogens in vivo. A growth chamber assay was performed to evaluate tomato plants response to strain Psf5. Walsh et al. [47] emphasized the need to investigate in situ colonization in the rhizosphere to determine the potential of any Pseudomonas strain as an effective BCA. Forty days after sowing of the inoculated seeds, Psf5 reached a population density of 106–107 CFU/g mixture under growth chamber conditions. In this system, strain Psf5 did not appear to negatively affect the development of tomato plants, but it also enhanced the growth of the root system. Several reports have indicated that IAA synthesis is related to plant growth stimulation by microorganisms, including P. putida [48,49]. IAA is the most common natural auxin found in plants and its positive effect on root growth and morphology is believed to increase the access to more nutrients in the soil. Thus, the production of IAA is a characteristic that may enhance Psf5 use as an effective biological control agent to contribute to the control of tomato damping-off caused by S. sclerotiorum.
In conclusion, the use of antagonistic bacteria fluorescent Pseudomonas as bioformulations for sustainable horticulture requires a thorough understanding of their functioning in the complex rhizosphere environment as well as of the response of vegetable crops to introduce microorganisms. Strain Psf5 showed in vitro inhibition of three fungal phytopathogens; it enhanced the growth of the tomato root system and it showed to be very promising in controlling tomato damping-off caused by S. sclerotiorum. Further work is underway in order to elucidate the specific factors involved in both growth stimulation and protection of tomato plants by Psf5. To conclude, a biocontrol system using the strain Psf5 may offer a good alternative to the currently employed chemical compounds, whose application entails a number of serious environmental and health hazards.
Disclosure of interest
The authors have not supplied their declaration of conflict of interest.
Acknowledgments
The authors wish to thank the Ministry of Agriculture and Water Resources and the Ministry of Higher Education and Scientific Research for the financial support of this research activity.


