1 Background
Lactoferrin (LF) is a member of the transferrin protein family, which is not only a highly abundant breast-milk glycoprotein [1,2], but, being produced by various mucosal epithelial cells, is commonly found in various secretory fluids, such as saliva, tears, and nasal secretions of different mammalian species, including humans, cows, goats, horses, dogs, and several rodents. Furthermore, LF has been identified in the eggs of rainbow trout indicating that this protein is also produced by fish [3]. LF is an essential player of the natural immunity. It is abundant in the upper and lower respiratory tissues, as well as in the digestive and urogenital mucosal tissues, which are often exposed to the colonizing or invasive microbial pathogens. It is broadly distributed in biological fluids, such as milk, saliva, and seminal fluid, and in specific neutrophil granules of polymorphonuclear leukocytes [4]. LF displays a broad range of preventive, therapeutic, and biological activities, including antibacterial, antifungal, antiviral, antioxidant, immunomodulatory, cell-growth modulatory, and binding functions, and neutralizes some bioactive substances, such as lipopolysaccharide (LPS) and glycosaminoglycan [5–9]. In the present work, we review the activities of LF directed against pathogenic viruses and, in particular, the mechanisms it uses against these pathogens.
2 Origin and structure of LF
Lactoferrin is a glycosylated globular protein first identified in 1939 in bovine milk [10], and subsequently separated from human milk [11]. LF is the essential iron-binding protein in milk, neutrophil granules, and exocrine secretions, such as tears, saliva, bile, pancreatic juice, small intestinal secretions, and cervical mucus. LF is a member of the transferrin protein family, with a net positive charge and pI in the range of 8.0–8.5 [12,13]. LF protein is expressed from a 35-kb gene conserved between different mammalian species (65–100% conservation) [12–15]. As shown in Fig. 1, LF is a single polypeptide chain, assembled into two symmetrical globular N- and C-terminal lobes [6]. Each lobe contains two domains, referred to as N1 and N2, or C1 and C2, which enclose a deep cleft in which the iron-binding site is located [16,17]. The two lobes are connected by a hinge region, which gives LF additional flexibility [1,18]. Each lobe can bind a single metal atom, such as the Fe2+ or Fe3+ ion, and also the Cu2+, Zn2+, and Mn2+ ions [12,13].
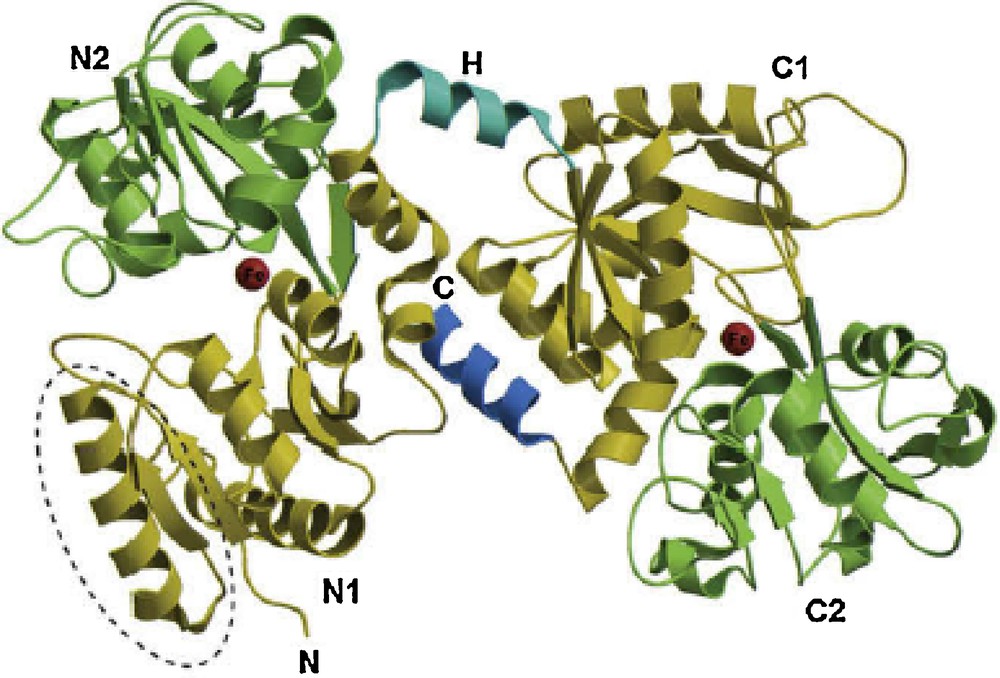
(Color online.) Structure of the holo form of camel LF. The N-lobe is on the left and the C-lobe is on the right, with the N- and C-termini of the polypeptide chain labeled N and C, respectively. The four domains are labeled N1, N2, C1, and C2, with the N1 and C1 domains shown in gold and the N2 and C2 domains shown in green. The connecting helix that joins the two lobes (H) and the C-terminal helix are shown in blue. The two iron-binding sites are shown as red spheres. The location of the antibacterial lactoferricin domain is outlined with a broken line.
The two lobes have internal amino acid homology of 40%, and it is presumed that during evolution, gene duplication resulted in the present LF gene (LTF) [14]. The active protein consists of about 690 residues [15,19].
2.1 N- and C-lobe structures
As other members of the transferrin protein family, LF consists of two homologous halves, the N- and C-lobes. The low level of iron saturation in natural LF and its excessively high binding affinity for iron allows it to insulate free iron from body secretions. This mechanism is harnessed by the LF found in neutrophils to prevent infection with pathogens, which are using this essential metal. Both LF lobes consist of about 345 residues, and the disposition of the two domains in each of the lobe forms an interdomain pocket (cleft) with high affinity for iron, binding of which is accompanied by synergistic binding of carbonate anions. LF has been shown to bind to different types of cells [20]. It also can bind to negatively charged molecules, such as DNA [21], heparin, other lipopolysaccharides (LPSs), and glycosaminoglycans [22]. This variable reactivity corresponds to the several suggested functions of this protein, which include the variety of roles, such as an antimicrobial agent, against both bacteria [23] and viruses [24,25], an antioxidant, a modulator of the immune and inflammatory responses [26,27], a growth regulator [28], transcription factor [29], regulator of the mitochondrial death signalling pathway [30], and an iron-absorbing agent [27].
The comprehensive analysis of the metal-unsaturated form of camel LF (cLF apo-form) crystal structure revealed that although the both lobes of this protein adopt open conformations, the disposition of amino acid residues in the iron-binding pockets of N- and C-lobs are dramatically different. The N-lobe of iron-free cLF has an amino acid configuration at the iron-binding pocket similar to that of the N-lobe of human apo-LF, whereas the residues of the iron pocket of the C-lobe are remarkably similar to that of hen and duck apo-ovotransferrins. Based on these observations, it has been concluded that the functional behavior of the N-lobe of cLF (binding and release of the iron ions) should be similar to that of human LF (hLF), whereas functionality of the cLF C-lobe resembles the functions of hen and duck ovotransferrins [31]. This hypothesis is in line with the remarkable difference in the pH-dependence of the iron release from the cLF N- and C-lobes, where the N-lobe of cLF loses its iron at acidic pH values (4.0–2.0), similar to the LFs of other species, whereas, similar to transferrins, the C-lobe of this LF loses its iron at higher pH values (7.0–6.0). Enzymatically produced N- and C-lobes of cLF also released their iron contents at pH 4.0 and 6.5, respectively. Therefore, cLF may have a dual functionality, with one half acting as LF and the other as transferrin [31].
2.2 Comparing LFs from different organisms
The LF sequences of nine species are currently known. These are LFs from human, bovine, camel, buffalo, sheep, goat, pig, horse, and mouse. It has been emphasized that there is a noticeable range in the LF pair-wise sequence identities [15]. This fact is illustrated by Fig. 2 where the pair-wise sequence identities of different LFs are shown. This analysis shows that the pair-wise sequence identities between Lfs from cow, buffalo, goat, and sheep exceed 90%, suggesting that they constitute a group of extremely closely related proteins. On the other hand, mouse LF, with its sequence identity in the 63–70% range, is the most different from other proteins. Next most divergent proteins are human and equine LFs, with the averaged sequence identities of 71.04% and 71.18%, respectively. Finally, pig and camel LFs have the averaged sequence identities of 71.78% and 74.25%, respectively (see Fig. 2). All this suggests that LFs represent a highly conserved family of proteins [15].
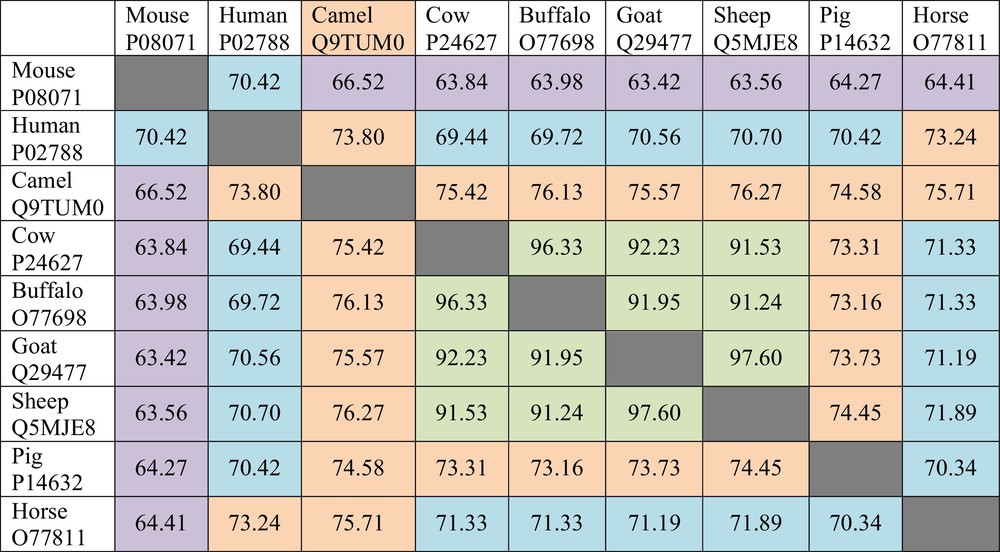
(Color online.) Pair-wise sequence identities of the lactoferrins from different species compared by the “Align” tool of the UniProt database (http://www.uniprot.org/align/).
With this level of sequence identity, one can expect that members of the LF family of proteins to be characterized by the significant structural similarity. This hypothesis is supported by the available crystal structures of the Lfs from human [32,33], cow [2], horse [34], buffalo [35], and camel [31]. The overall structural organization of these proteins is well preserved, with all of them being folded into two globular lobes, each of which is further subdivided into two α/β domains consisting of a central, mostly parallel β-sheet, with α-helices packed against it. These α/β domains are known as N1 and N2, or C1 and C2, and in each lobe, these domains enclose a deep metal-binding cleft [15]. Such structural organization of each lobe represents two-domain “binding protein fold”, which can be found in bacterial periplasmic proteins that transport ions and small molecules, suggesting common evolutionary origin of thee bacterial transporters and LFs [15,36].
Despite the overall structural similarity between different lactoferrins and their high sequence identities, it has been noted that the N-terminal part of cLF (first 50 residues) is very divergent from the corresponding regions of other LFs [31]. In fact, although pair-wise alignments of the full-length LFs revealed that cLF is ∼ 74% identical to other members of this family, the identity of its N-terminal tail is essentially lowers, being less than 40% [31]. It was also pointed out that dynamic properties of cLF can be distinct from those of other LFs, since cLF is different from other LFs in several critical residues related to the domain movements associated with the metal-binding (e.g., residues Pro418, Leu423, Lys433, Gln561, Gly629, Lys637, Arg652 and Pro592) [31].
Importantly, lobes in buffalo, cow, and camel LFs are connected by a hinge region which is specifically attacked by a proteinase K that cuts these proteins at the interlobe connecting peptide, generating functional N- and C-terminal half-molecules [15,31,37–39]. It was also pointed that that the complex two lobes, four-domain structure defines the dynamic properties of Lfs and explains the ability of these proteins to undergo large-scale rigid-body domain movements associated with metal-binding, where binding and release of metal require closing and opening of the two domains of each lobe [40,41]. Although the Lfs of different species differ only very little in the extent to which their domains close over a bound metal ion, there is a noticeable divergence between the Lfs of different species in the relative orientations of their N- and C-lobes [15]. This structural variability dictates the difference in the interlobe contacts in different LFs and also regulates the extent or nature of cooperativity between the lobes [15]. Curiously, of all crystal structures of metal-depleted LFs from different organisms, apo-cLF was shown to be characterized by the most open conformation of both metal-binding clefts, where the N2 and C2 domains move away from the N1 and C1 domains, respectively, and where the open iron-binding clefts in the N- and C-lobes are rotated with respect to each other by 67.1° [31].
2.3 Intrinsically disordered regions in lactoferrin
Many of the functional features of lactoferrin, including its multifunctionality, metal-binding capability, presence of multiple post-translational modifications (such as phosphorylation, acetylation, and glycosylation), and the ability to be present in at least two isoforms due to the alternative promoter usage are typical characteristics of intrinsically disordered proteins or hybrid proteins possessing ordered domains and disordered regions [42–53]. In fact, such proteins are common in various proteomes, where they play numerous biologically important roles, being predominantly involved in cell signalling, and participating in regulation and control of various cellular processes [53–60]. Disordered and hybrid proteins, being pliable and highly adjustable, have multiple functional advantages over the ordered proteins [45,46,53,60,61]. Among these advantages are the ability to be involved in multiple binding events and to fold differently at interaction with different binding partners [50,62]. Disordered and hybrid proteins are also very common in various human diseases [46,63–69], and are abundantly present in viruses [58,70,55,71–75] and pathological microbes [76,77]. Furthermore, intrinsic disorder is important for functionality of proteins involved in the innate immune response [78].
To visualize the intrinsic disorder status of hLF, we used the graphing facilities of the D2P2 database (http://d2p2.pro/) [79]. Fig. 3 represents results of this analysis and shows that hLF, being mostly ordered still possesses numerous disordered regions. In agreement with earlier studies, many of the sites of post-translational modifications are located within or in close proximity to these disordered regions. Of special importance is the N-terminally located disordered region (residues 38–59) that corresponds to the LF-derived cell-penetrating peptide (CPP) with the conformation-dependent uptake efficiency [80] and which is a part of the lactoferricin-H molecule. In agreement with these computational predictions, Duchardt et al. [80] showed that this 22-residue-long CPP, which corresponds to the loop and one disulfide bridge of human lactoferricin, is predominantly disordered in its unbound form irrespectively of the status of the disulfide bridge (i.e., both oxidized and reduced forms of this CPP were equally disordered), but undergoes a profound transition to the β-sheet structure upon interaction with the large unilamellar vesicles of different composition [80]. The efficiency of interaction and degree of structural transformation were shown to be dependent on the oxidation state of this peptide [80].
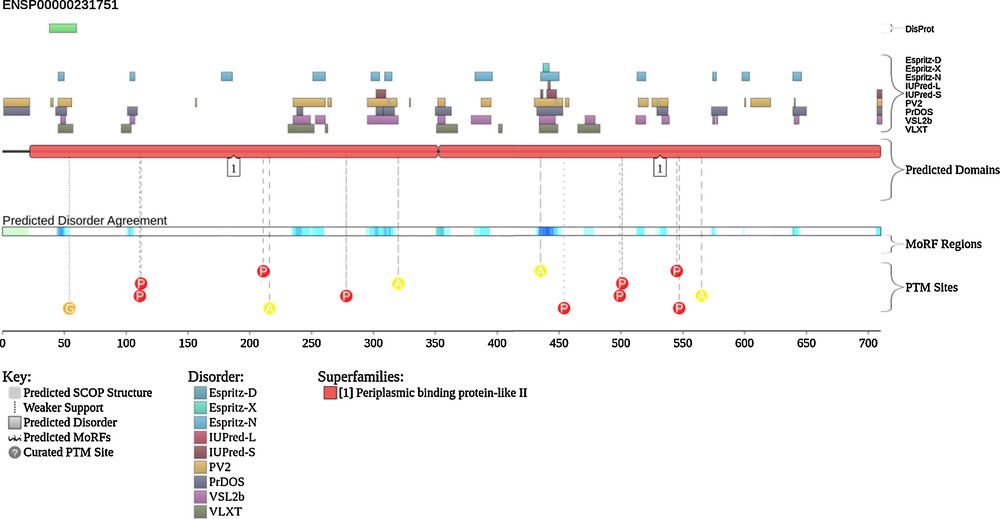
(Color online.) Intrinsic disorder status of lactoferrin visualized using the D2P2 database (http://d2p2.pro/). The amino acid sequence of human protein (UniProt ID: P02788) was used in this analysis. Keys to the outputs of various computational tools are shown in this plot.
Also in line with these observations for human lactoferricin, bovine lactoferricin B, a 25-residue peptide excised through pepsin cleavage in the stomach from the intact bLF, was shown to possess remarkably different structures in solution and in crystal. In fact, this region in crystal structure of the intact bLF represents an α-helix followed by a single β-strand [2] (see Fig. 1), whereas in the NMR experiments, this peptide displayed a slightly twisted anti-parallel β-hairpin structure [81]. Furthermore, careful molecular dynamic simulation analysis of bovine lactoferricin B, either starting from NMR-determined solution structure or from the structure of this region excised out of the intact protein, revealed that although the NMR-based β-sheet structure was relatively stable, the peptide released from the protein, being relatively unstable, melts rather fast and can converge to the structure similar to the NMR solution structure under the effect of small directional force [82]. Since the mentioned region of the protein can exist in at least three different conformations (disordered, β-hairpin, and an α-helix followed by a β-strand), this region of the lactoferrin clearly belongs to the category of the chameleon sequences [83–86], the ability of which to change their structure depending on the peculiarities of their environment is determined by their high intrinsic disorder propensities.
3 Production of natural and recombinant LF
The scaling-up of experimental protocols for industrial production of LFs is a quite difficult task since there are several obstacles in the manufacturing of pure LF. These obstacles in LF mass production include the requirement for the reuse of residual milk materials after the LF purification, the use of a simple manufacturing process, the product stability, and the high levels of sanitation required for plant equipment. However, because LF has many functions and uses, attempts have been made to develop commercial production strategies to produce recombinant LF or to purify the protein for use as a food additive or therapeutic agent. LF purification strategies depend on the physical and structural characteristics of the protein, such as its net positive charge, its ability to bind iron, and its glycosylation status [87,88]. Because LF has a characteristic positive net charge, it is efficiently adsorbed onto cationic exchange resins and is easily released from the resin with saline solutions. This protocol was efficiently employed for the purification of human, bovine and camel LFs (hLF, bLF, and cLF, respectively) [24,87,89–91]. Because LF binds Fe3+, it can be purified by metal ion affinity chromatography [24]. Based on its glycosylation pattern, LF can also be purified downstream by affinity chromatography, for instance on concanavalin A Sepharose or heparin Sepharose [90,92]. The production of LF by its isolation and purification from milk has a number of potential safety problems, such as contamination with viral and/or bacterial pathogens, and can also be limited by the available supply of milk. The need for greater amounts of LF has led to the development of strategies to produce a recombinant form of LF (rLF). Many competing approaches to the production of rLF (Table 1) have involved the production of this protein by bacteria, fungi, insect cells, plants, and transgenic animals [93]. Baby hamster kidney cells were the first expression system used to express recombinant hLF [94]. Numerous expression vectors (Table 1) have been developed for yeasts, bacteria, and plants, and have produced full-length rLF, as well as individual LF lobes and LF-derived peptides. Viral vectors have also allowed the expression of rLF in insect cells, in cell culture, or in organisms, in which recombinant hLF, bLF, and cLF have been expressed. The expression of rLF in higher eukaryotic organisms has been achieved with microinjection, and later with the transfection of viral vectors into the mammary gland, creating transgenic animals, such as goats, mice, rabbits, and cows, that produce milk containing rLF (Table 1) at concentrations up to 2 g/L (as in the milk of transgenic goat). Higher plants can also synthesize correctly folded LF with the appropriate levels and patterns of glycosylation and with normal functional activity. Recombinant hLF and bLF have been successfully expressed in plant systems (Table 1). A recent review by Yemets et al. [95] provides a detailed analysis of the advantages and disadvantages of the existing non-animal systems for the LF expression, with the special focus on using higher plants as bioreactors for the large-scale LF production. The authors describe protein yields and biological activities of LF produced in different systems and emphasize the benefits of monocot plant system for LF expression [95]. Curiously, the expression of LF in plant models could significantly increase crop quality by improving their resistance to some plant pathogens. It can also provide a source of high-quality recombinant protein [96]. Most expression systems that have been developed to produce rLF or its derivative peptides generate proteins with physical, biochemical, and biological properties very similar to those of the native form, including their molecular mass, glycosylation, antimicrobial and anti-inflammatory activities, thermostability, and capacity to bind iron. These expression systems are important for the economic, high-scale production of rLF [97].
Expression of rLF and its derivatives by various transgenic organisms.
| Protein/peptide | Expression system | LF origin | Expression concentration | Reference |
| Full-length lactoferrin | ||||
| Bacteria | ||||
| E. coli | kLF | 17 mg/L | [113] | |
| Lactobacillus casei | hLF | 10.6 mg/mL | [114] | |
| Yeast | ||||
| Pichia pastoris | hLF | 115 mg/L | [115] | |
| capLF | 2 mg/L | [116] | ||
| pLF | 12 mg/L | [117] | ||
| yLF | 40 mg/L | [118] | ||
| eLF | 40 mg/L | [119] | ||
| hLF | 1200 mg/L | [120] | ||
| Pichia methanolica | pLF | NR | [121] | |
| Fungi | ||||
| Aspergillus awamori | hLF | 2 g/L | [122] | |
| Aspergillus oryzae | hLF | 25 g/L | [123] | |
| Insects cell lines | ||||
| Spodoptera frugiperda | cLF | 2 mg/L | [36] | |
| hLF | 9.5 mg/L | [124] | ||
| Bombyx mori | hLF | 65 mg/L | [125] | |
| hLF | 13.5 μg/1–2 × 105 cells | [126] | ||
| pLF | 205 μg/pupa | [127] | ||
| Cell lines | ||||
| BHK | hLF | 20 mg/L | [38] | |
| Cell culture | ||||
| Human embryo kidney | hLF | 16 mg/L | [128] | |
| Mammals | ||||
| Goat | hLF | 0.756 mg/L | [129] | |
| hLF | 2 g/L | [130] | ||
| Mice | hLF | 2.5 mg/mL | [34] | |
| hLF | 0.2–2.5 g/mL | [131] | ||
| Rabbit | hLF | 2.3 mg/mL | [132] | |
| Bovine | hLF | 1 g/L | [133] | |
| hLF | 2.9 mg/mL | [134] | ||
| Plants | ||||
| Nicotiana tabacum | hLF | NR | [135] | |
| hLF | 0.1–0.8% SP | [124] | ||
| hLF | 4.3% SP | [136] | ||
| bLF | NR | [137] | ||
| Rice | hLF | 1.6 mg/g seed | [138] | |
| hLF | 0.5% total biomass | [139] | ||
| hLF | 2–4% SP | [140] | ||
| Potato | hLF | 0.1% SP | [141] | |
| Sweet potato (Ipomoea batata) |
hLF | 3.2 μg/mg total protein | [142] | |
| Ginseng (Panax ginseng) |
hLF | 3.0% SP | [143] | |
| Tomato (Lycopersicon esculentum) |
hLF | NR | [144] | |
| Maize | hLF | NR | [145] | |
| Alfalfa (Medicago sativa) | hLF | NR | [146] | |
| Barley | hLF | NR | [147] | |
| Lactoferricin | ||||
| Bacteria | ||||
| E. coli | bLFc | 10 mg/L | [148] | |
| LFc | 60 mg/L | [149] | ||
| bLFc | 2 mg/L | [150] | ||
| Yeast | ||||
| Pichia methanolica | bLFc | 90 mg/L | [151] | |
| N- and C- lobes of LF | ||||
| Bacteria | ||||
| Rhodococcus erythropolis | bLF C-lobe | 3.6 mg/mL | [149] | |
| Insects cell lines | ||||
| Spodoptera frugiperda | cLF N-lobe | 2 mg/L | [36] | |
| cLF C-lobe | 2 mg/L | [36] | ||
| Plants | ||||
| Nicotiana benthamiana | hLf N-lobe | 0.6% SP | [152] |
4 Antimicrobial activities of LF
Lactoferrin is a crucial constituent of the innate immune system on mucosal surfaces, where it has several functions and can respond to a variety of physiological and environmental factors [98]. LF exerts antimicrobial activity against many different infectious microbes. This activity is mainly attributable to the ability of this protein to sequester iron with high affinity and to retain the bound iron under acidic conditions. However, the structural features of LF may provide this protein with additional functions that are not restricted to maintaining iron homeostasis. In agreement with this hypothesis, it has become clear that some of the antimicrobial characteristics of LF are independent of its iron-binding mechanism [99–101] and are attributed to direct interference of LF with the bacterial cell surface [99,102,103]. LF exerts distinctive antimicrobial activities against a broad range of Gram-positive bacteria, Gram-negative bacteria, fungi, yeasts, viruses [5,8,9], and parasites [104].
Curiously, LF does not only have profound antimicrobial activity against pathogenic microbes, but was shown to promote the growth of beneficial bacteria, such as those belonging to the Lactobacillus and Bifidobacteria genera [105]. The precise mechanisms of these stimulatory effects of LF on beneficial bacteria are poorly understood. Since treatments with recombinant hLF (rhLF) alone, Lactobacillus casei sps. rhamnosus (LGG) alone or rhLF + LGG combined were shown to enhance neonatal intestinal host defences and alter the colonization or infection of the small bowel with E. coli, and since the levels of prophylactic rhLF introduced into the gut before infection were far too low to hinder the intestinal growth of E. coli via an iron withholding, the authors proposed that the beneficial actions of LF at epithelial surfaces could be attributed to the immunostimulatory and/or immunomodulatory effects of this protein [105].
5 Antiviral activities of lactoferrin
The advantages of breast-milk are not restricted to its nutritional value, but include its many distinctive antiviral components. However, most of the antiviral activities attributed to milk can be ascribed to LF alone, and Fig. 4 shows that this protein possesses a broad antiviral capacity against very different viruses. The antiviral activities of hLF were first described in mice inoculated with a polycythemia-inducing strain of the Friend virus complex (FVC-P) [106]. Since 1994, the antiviral activities of LF against both naked and enveloped human and animal pathogenic viruses have been explored.
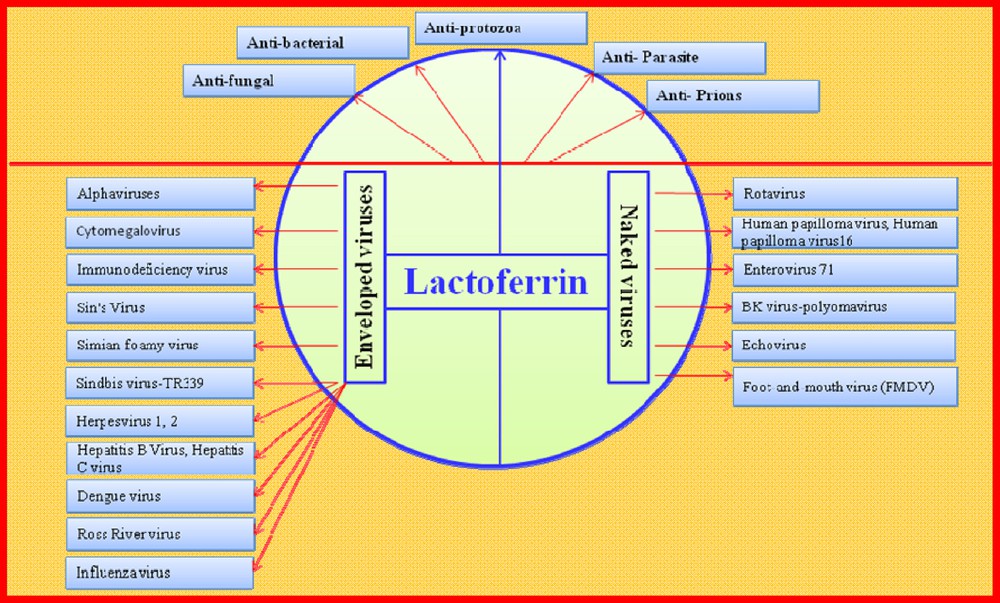
(Color online.) Multiple antimicrobial functions of lactoferrin. The antibacterial activities of lactoferrin against the Gram-positive and negative bacteria, and the antiviral activities of lactoferrin against the non-enveloped (naked) and enveloped viruses are shown. Viral abbreviations: HCMV (human Cytomegalovirus), HIV (human immunodeficiency virus), SIN (Sindbis virus), SFV (Simple file verification), TR339 (Sindbis virus (SINV) strain TR339), TRSB (Sindbis virus strain TRSB), HSV-1, HSV-2 (herpes simplex virus type 1) (HSV-1) and type-2 (HSV-2), HCV (hepatitis C virus), HBV (hepatitis B virus), Ross River V (Ross River virus) (RRV), HPV (Human papillomavirus), HPV-16 (Human papillomavirus-type 16), EV71 (Enterovirus 71), BK-polyomavirus (polyomavirus), Foot-and-mouth virus (Foot-and-mouth disease virus, FMDV).
5.1 Molecular mechanisms of the LF antiviral activity
A panel of experimental bioassays has been developed to study the mode of action of LF against several viruses. Since the antiviral activity of LF may be exerted by interfering with a cellular target, the LF activity is screened by pre-incubating cells with the protein before they are infected with viral particles, e.g., hepatitis B virus (HBV), HS-adapted Sindbis virus, Semliki Forest virus, CMV, HSV-1, and HSV-2 [107–110]. In contrast, LF can also exert its antiviral activity by interfering directly with viral particles, e.g., adenovirus, feline herpes virus (FHV-1), HCV, and HIV [89,91,92,111–114]. In many reported cases, LF seems to exert its antiviral activity in an early phase of the infection process [108–110,112,113]. Recently, similar results have been reported with cLF, when the pre-incubation of HCV with cLF totally prevented viral entry, whereas when human leukocytes [89], HepG2 cells [91], or Huh7.5 cells [92] were pre-incubated with LF before HCV infection, there was no effect on the viral entry. Camel LF can inhibit the intracellular replication of HCV [91,92,111]. Many viral pathogens use the host cell surface heparan sulfate pathway of receptor attachment during the infection process. LF has also been reported to bind to heparan sulfate [115], and several studies have demonstrated that heparan sulfate at the cell surface is important for the antiviral activity of LF [109,116]. Curiously, a recent study by Calvarho et al. [117] demonstrated that the sulfate moiety of this glycosaminoglycan is responsible for the antiviral activity of LF, since the efficiency binding of bLF to the cell was highly dependent on the sulfation degree of glycosaminoglycans [117].
In an attempt to understand the molecular mechanisms of the antiviral hLF activity, Florian et al. [118] characterized the capacity of this protein and the hLF-derived synthetic peptides (HLP) corresponding to the N-terminal region of this protein (1–47 amino acids sequence) to prevent hepatitis B virus (HBV) infection and replication. The authors used the HepaRG and HepG2.2.2.15 cell lines to show that some of these peptides possess efficient (40–75%) inhibition of HBV infection [118]. The most active was the HLP1-23 peptide containing the GRRRR cationic cluster, which corresponds to one of the two glycosaminoglycan binding sites of hLF. Since pre-incubation of this peptide with the virus was able to inhibit the HBV infection, and since no effect was observed on the target cells, the authors concluded that the cationic GRRRR cluster was sufficient for the stable binding of this peptide to the negatively charged residues on the virion envelope, whereas the efficient attachment of the HLP1-23 peptide to the cells was prevented due to the absence of the second glycosaminoglycan binding site [118].
The mode of action of LF is not restricted to the direct or indirect interactions with viral molecules and/or host cells, but may be attributed to its immunomodulatory capacity [119]. Interferon α (IFN-α) is used as a drug to treat HCV infection. Tablet-form, liposomal, and free LF have been shown to induce and regulate IFN-α production in healthy volunteers [120]. LF encased in multilamellar liposomes has significantly suppressed CCL4- and LPS-induced tumor necrosis factor α (TNF-α) hepatotoxicity in the rat liver [120]. However, pre-treatment of these injured rodent livers with polyethylene-glycosylated (PEGylated) LF significantly reduced hepatocyte damage. PEGylated LF exerted a significantly greater hepatoprotective effect than free LF [121]. It also improved several lipid-metabolism-related parameters [122]. The oral administration of LF to a murine model of hepatitis enhanced the activity of circulating natural killer lymphocytes [8], suppressed hepatic inflammation, and induced the expression of interleukin-11 (IL-11) [123]. The continuous intake of a dietary supplement containing LF by experimental piglets increased their immunostimulatory capacity and improved their immune responses [124,125].
5.2 Lactoferrin activity against hepatitis C virus (HCV)
As follows from Fig. 4, LF has very prominent antiviral activity against a very broad spectrum of enveloped and naked viruses. This section is dedicated to the description of the LF activity against hepatitis C virus (HCV). This section is a bit more detailed than the subsequent description of the activity of LF against other viruses. This is explained in part by the fact that the related research is among the main interest of the authors. Also, the anti-HCV activity of cLF has been relatively well studied, and the treatment of HCV with cLF has reached clinical stages (see below).
Hepatitis C virus (HCV) is responsible for a blood-borne disease causing hepatocellular injury, which is accompanied by numerous immunological events. HCV is considered the most significant public health problem in Egypt, with infection prevalence of up to 20%. This is 10 times greater than in any other country in the world. The highest genotype prevalence of HCV is genotype 4, which is responsible for about 90% of infections, with the predominant subtype being 4a (55%). Although HCV is estimated to infect 180 million individuals worldwide, and is the leading cause of liver-associated disease, this virus still lacks any effective therapeutic drug or preventive vaccine [126,127], and many infected patients resort to the sparse knowledge available on alternative medical strategies to cure themselves. LF is considered as one of those alternative medicinal agents, displaying noticeable antiviral activity against HCV in vitro and in vivo. LF inhibits the entry of HCV into the human hepatocyte cell line PH5CH8 [128]. The pre-incubation of LF and HCV particles prevents the attachment of the virus to its cellular receptors and consequently inhibits viral infection. It is hypothesized that LF prevents HCV infection by directly interfering with the viral particle. Ikeda et al. [129] have shown that the domain responsible for the antiviral activity of LF against HCV does not necessarily correspond to its bactericidal domain.
Bovine and human LFs can attach to HCV envelope proteins E1 and E2. This attachment interrupts the interaction between the viral particle and its cellular receptor CD81. Nozaki et al. [130] have shown that the peptide comprising the 93 amino acids at the C-terminus of LF is partially homologous to CD81. They have also shown that the antiviral properties of this 93-residue peptide are actually confined to a segment of only 33 amino acids. Heat-treated LF shows no antiviral activity against the virus, indicating that the natural conformation of LF is required for its antiviral effect. Lactoferricin can also prevent HCV entry into cells [129]. Reducing the incubation times of LF with HCV enhances the rate of viral infection. On the contrary, increasing the incubation times probably allows sufficient time for LF to interfere with the adsorption of HCV onto its target cells [128,129]. Yi et al. [131] showed that LF binds the HCV E2 glycoprotein, thus interfering with cell infection, because an anti-hLF antibody co-precipitated both the secreted and intracellularly expressed forms of E2. Native and recombinant cLF also inhibited the cellular entry of HCV when LF and the viral particles were pre-incubated together in vitro [89,91,92]. The activities of the recombinant and native versions of cLF did not differ significantly [92], which may reflect the identical structural properties functional antiviral potential of these two forms of LF. It seems that the carbohydrate moieties have no role in the anti-HCV activity of cLF. However, the N- and C-lobes of cLF, separated with proteinase K, both displayed significant antiviral activity against HCV [92]. Recently, camel LF and its N-lobe were produced in a baculovirus expression system. The anti-HCV activity of the recombinant N-lobe of cLF was 2–3-fold higher to that of the native full-length LF or its fragments [92]. A recent study that investigated different types of LF purified from human, camel, sheep, and goat milk revealed that all the LFs effectively inhibited HCV and prevented HCV entry into HepG2 cells and/or inhibited its intracellular replication. Camel LF had the highest antiviral activity in comparison with other LFs [111], similar to the previously reported antibacterial activity of this protein [132].
When four camel milk proteins (LF, amylase, immunoglobulin G [IgG], and casein) were screened, they showed anti-HCV activities ranging from undetectable (amylase, casein) to intermediate (IgG) to strong (LF) [89,91,92,111]. In another study, we found that casein and α-lactalbumin failed to inhibit the entry of HCV into peripheral blood mononuclear cells or Huh7.5 cells [133]. Furthermore, in a recent study evaluating the beneficial effects of raw camel milk on the general health and fitness of HCV patients, we found that raw camel milk reduced the HCV RNA load in more than 80% patients. The chronic HCV patients thus treated showed antibody-isotope profiles skewed toward Th1 immunity (manuscript in preparation). We also found that herbal medicines, such as laccase, which was extracted from the oyster mushroom (Pleurotus ostreatus), showed antiviral activity against HCV and could be used as a natural and alternative medicament to control HCV infection, specifically in patients in the chronic phase [134]. However, cLF has proved to be more efficient than mushroom laccase.
The synthetic peptide Cs3–33 (amino acids 600–632) derived from the C-lobe of hLF specifically inhibited the entry of HCV into PH5CH8 cells by binding to HCV envelope protein 2 (E2), but it was less active than intact LF [129,135]. However, tandem repeats of this peptide sequence (Cs3–33 × 2 or × 3) demonstrated stronger anti-HCV activity in the same cell line; the triplicated peptide was stronger than the duplicated peptide, and both of them were stronger than the single peptide [135]. The results were similar when a vesicular stomatitis virus pseudotype carrying the green fluorescent protein (GFP) and native E1 and E2 genes was used [136]. Beleid et al. [137] evaluated the activity of five synthetic peptides with different numbers of residues (17–33 residues from the region defined by amino acids 600–632) from the human LF C-lobe against HCV cellular entry. These peptides have varying levels of helical content. Two of the five peptides specifically bound viral E2 and prevented HCV cell entry. Surprisingly, these two peptides had the greatest amount of helical structure, which probably means that the peptide binding affinity for the viral envelope protein increases as the peptide helicity increases [137]. In contrast to the Nozaki peptide (from the C-lobe of hLF), which was weakly bound to the HCV E2 peptide, the cyanovirin N-peptide (101 residues) was specifically bound to the HCV E2 glycans and inhibited viral cell entry by preventing the HCV E2–CD81 interaction [130]. Currently, a small peptide from hLF is undergoing phase II clinical trials as an antimicrobial drug for allogenic bone-marrow stem-cell transplantation [138].
5.3 LF activity against herpes simplex viruses
Iron-saturated LF has been shown to exert antiviral activity against herpes simplex virus-1 (HSV-1) and HSV-2 [100,139–141]. Antiviral potential has been ascribed to both apo- and holo-LF [140], which means that the antiviral activity of LF does not depend on the presence of iron ions in the molecule. It is important to remember though that although this observation is true for most of the virus species tested, for a few virus species, such as HIV (see below), apo-LF presents an antiviral activity greater than holo-LF.
The adsorption of HSV to its heparan sulfate cellular receptors is blocked by LF [139]. The authors of that study suggested that this block may be ascribable to the N-terminal residues of LF, which compete with the virus for the cellular receptor. Siciliano et al. [142] also demonstrated that the bLF anti-HSV activity is mediated specifically by its N-lobe (amino acids 1–280), although the C-lobe (amino acids 345–689) also displays variable activity, but it is weaker than that of the N-lobe. These authors also demonstrated that HSV-1 was prevented from infecting cells by synthetic peptides from the LF N-lobe (amino acids 222–230 and 264–269) [142]. Shorter synthetic peptides, such as bovine lactoferricin, also ameliorated HSV-1 movement from cell to cell [143]. Hammer et al. [144] showed that lactoferricin, a 24-residue peptide derived from the N-lobe, exerts antiviral effects against HSV. The studies of both Siciliano et al. [142] and Hammer et al. [144] demonstrated that the antiviral effects of native LF are more potent against HSV than are those of synthetic peptides.
5.4 Lactoferrin and cytomegalovirus (CMV)
The antiviral effects of both bLF and hLF against cytomegalovirus (CMV) have been reported [107]. Total cell protection was achieved when LF and CMV were co-incubated before they were mixed with the host cells. This cell protection possibly occurred because of the direct interaction between LF and the virus. However, a possibility of the LF molecule attachment to the cellular receptors of CMV could not be excluded. Harmsen et al. [145] showed that the amidated LF has greater antiviral potency against human CMV (HCMV) than the succinylated form of LF. These authors also demonstrated that almost complete protection was achieved when fibroblast and lymphocyte cell lines were pre-incubated with LF for 30 min before exposure to HCMV. The N-terminal region of LF seemed to be crucial for its antiviral activity. LF has also shown a protective effect in vivo in a murine model of the HCMV infection. Here, animals treated with LF survived challenge with a lethal dose of mouse CMV, after the antiviral activity of LF was optimized at the start of treatment and before infection [25].
5.5 Lactoferrin as an anti-HIV agent
Bovine and human LFs are potent inhibitors of human immunodeficiency virus (HIV) infection in vitro [145–147]. Harmsen et al. [145] demonstrated that among many proteins evaluated, LF was the only protein to prevent HIV-1 replication inside MT4 cells. Both Harmsen et al. [145] and Puddu et al. [146] showed that the mechanism of LF action against HIV involves the interruption by LF of the adsorption of the virus to its target cells in an early phase of infection. LF can bind to the GPGRAF domain of the gp120 glycoprotein of HIV [147]. It is possible that the binding of LF to gp120 is responsible for its antiviral effects against HIV, because the binding of gp120 to the CD4 or chemokine receptors on the target cells plays an important role in the adsorption and entry of HIV into those cells [148–150]. Legrand et al. [151] and Groot et al. [152] have also demonstrated that LF can bind to three HIV co-receptors and to the DC-SIGN receptor. Furthermore, the binding of LF to surface nucleolin was shown to block the attachment and entry of HIV molecules into HeLa P4 cells [151]. These studies showed that both apo-LF (metal-unsaturated LF) and holo-LF (iron-saturated LF) display antiviral activity against HIV, although apo-LF is more potent than holo-LF [151].
5.6 Lactoferrin activity against the simian rotavirus
LF has prominent activity against the simian rotavirus SA11 in vitro [152]. The mechanism of action of LF against this rotavirus involves its binding to the viral particle, thus inhibiting the adsorption of the virus to the target cell [152]. Although Superti et al. [153] demonstrated that this antiviral effect is associated with the N-lobe, they suggested that LF does not inhibit viral attachment to the cellular receptors by competing for common binding sites on the cells, because the SA11 rotavirus receptor is glycosylated differently from glycosaminoglycans. Furthermore, flow cytometry revealed an interaction between LF and the viral particles. LF also inhibits rotavirus intracellularly by withholding calcium, which is important for the assembly of new viral particles [152]. However, LF saturated with different metal ions also has antirotaviral activity [152]. The antirotaviral activities of LF saturated with different metals ions are very similar, and both apo- and holo-LF can bind to the viral particles and prevent viral attachment [152,153].
5.7 Lactoferrin and poliovirus
LF has also shown antiviral activity in vitro against poliovirus [154]. Both apo-LF and LF saturated with different ions (Fe3+, Zn2+, and Mn2+) can prevent poliovirus infection [154]. LF acts against poliovirus in the acute phase of infection, using a mechanism that interrupts the entry of the virus into the target cell [154].
5.8 Anti-adenovirus potential of LF
Human and bovine LFs have antiviral activity against adenoviruses [155]. Bovine LF prevented an early step in EV71 viral infection when premixed with the epithelial cells or when added during the viral adhesion step, and prevented adenoviral antigen synthesis. The antiviral activity of LF against adenoviruses is mediated by the N-lobe, and the C-lobe has no inhibitory effect [156]. Arnold et al. [155] and Di Biase et al. [156] demonstrated that lactoferricin alone is sufficient to inhibit adenoviral entry. The mechanism of action of LF against adenovirus is exerted during the virus attachment step and LF also inhibits virus replication, mainly by competing for the cellular membrane glycosaminoglycan. LF also interferes with the adenoviral infection by binding to the viral envelope structures, specifically to the adenovirus penton base (polypeptide III), the protein responsible for the attachment of the virion to the cell receptor integrins and its subsequent internalization [157]. LF also influences the levels of inflammatory mediators by activating natural killer cells, indirectly contributing to the inactivation of microbes in neutrophils [158].
5.9 LF activity against Mayaro virus (MAYV)
Recently, Calvarho et al. [117] reported the prominent antiviral activity of bLF against an arbovirus, Mayaro virus (MAYV), causing a highly debilitating febrile illness in many regions of South America, with no effective antiviral intervention being available against human infections with MAYV. Careful analysis of the apo-bLF effects on the different stages of MAVY infection of baby hamster kidney (BHK-21) and African green monkey kidney (Vero) cells revealed that the most compromised stage is the virus entry into the target cell [117]. The bLF-controlled inhibition of virus entry to the target cell was shown to be dependent on cellular glycosaminoglycans [117]. In fact, the authors showed that since the sulfated glycosaminoglycans on the cell membrane were needed for the Mayaro virus entry, the adhesion of virus to host cells was impaired by the bLF-dependent blockage of glycosaminoglycans [117]. Curiously, Calvarho et al. [117] also showed that apo-bLF promoted a slight increase in viability of control, virus-untreated cell. This increase in viability was promoted by a reduction in cell death by apoptosis that might occur as a result of stress coming from the cell confluence in culture [117].
5.10 In vivo animal models to study the antiviral activities of LF
Although LF clearly shows very prominent antiviral activity against various viruses in vitro, careful analysis of the in vivo animal models is needed to produce conclusive evidence on the suitability of this protein as potential antiviral drug. In a recent study, Gualdi et al. [159] showed that there is a contradiction between the results of in vitro and in vivo experiments. Here, despite the fact that LF was shown to have antiviral activity against respiratory syncytial virus (RSV) in vitro, an in vivo murine model, bLF administered by different routes [by oral gavage or intraperitoneal (IP) injection] and at different doses did not demonstrate anti-RSV activity in mice [159]. These observations suggest that care should be taken while transferring the in vitro results to the in vivo models.
6 Lactoferrin and adaptive immunity
Lactoferrin has a unique immunomodulatory action on adaptive cellular functions, on both T and B lymphocytes and other immune cells, by promoting the maturation of T-cell precursors into competent helper cells and the differentiation of immature B-cells into efficient antigen-presenting cells (APCs) [124,125,160]. The relationship between the cellular and humoral immune responses is strong, and many cytokines and mediators have been identified as early mediators of the effector T-cell development [161]. Recent studies have found evidence that LF exerts immunoregulatory effects on Th1/Th2 cell activities. LF can stimulate the differentiation of T-cells from their immature precursors by inducing the expression of the CD4 antigen [162].
Most studies have focused predominantly on the relationship between LF and CD4+ T-cells and the roles of LF in disease by affecting the Th1/Th2 cytokine balance in a manner that is dependent on the host's immune response. LF causes a Th1 polarization in diseases in which the ability to control infection or tumor relies on a strong Th1 response [162]. Th1 cells stimulate and activate macrophages, resulting in intracellular killing events that eliminate intracellular pathogens, often through the production of reactive oxygen intermediates [163]. Mincheva-Nilsson et al. [164] showed that LF receptors are expressed on all T-cell subsets, including δγ T-cells. LF can also bind to human T-cell surface receptors [165,166], and LF binding may be involved in receptor-mediated endocytosis [167]. Both Constant and Bottomly [168] and Sanders [169] reported that the Th2 subtype stimulates and activates B-cells, allowing isotype switching and promoting the production of antigen-specific antibodies. Wang et al. [170] reported that LF elevated the lymphoid and resident intestinal CD4+ and CD8+ T-cells in mice implanted with tumor cells.
LF modulates T-cell activity and their reactivity to local antigens [163]. LF can induce cellular recruitment, causing APC activation, and thus enhancing antigen-specific immune responses. Therefore, LF is considered a bona fide alarmin that favors a Th1-type immune response [171]. The effect of LF on T-cell populations can be further defined as specifically targeting a subset of cellular responses. A well-studied phenomenon is the ability of LF to modulate and directly change the balance between the Th1 and Th2 immune responses, often defined by the T-cell cytokines IFN-γ and IL-4/IL-5, respectively. As with the overall T-cell response, LF affects both Th1 and Th2 cellular subsets. In addition to the role of LF as a modulator of excessive responses, it can also promote the maturation of B-cells by increasing the expression of complement 3 receptor (C3R), promoting the acquisition of surface IgD, and enhancing B-cell antigen presentation [160].
The oral consumption of LF by chronically viremic HCV patients not only promotes the inflammatory cascade and recruits and activates APCs, but also potentiates the production of Th1 cytokines in the peripheral blood, specifically IL-18 [145]. The ingestion of lactoferrin professionally stimulates the gut-associated immune response through the expression of IL-18 and type I IFNs, in addition to increasing the activity of δγ T-cells and natural killer lymphocytes [132]. Therefore, depending on the infectious phases and viral burdens of HCV patients, LF may promote their T-lymphocytes towards either a Th1 or a Th2 immune response [172,173].
7 Conclusions
Lactoferrin has become a very well-known antimicrobial protein since it was first isolated and characterized, although the mechanisms of its antimicrobial activity are still under investigation. Although the antimicrobial activity of LF against bacteria was investigated first, its activity against other microorganisms, such as viruses, fungi, and parasites, was subsequently recognized. Concerns about the appearance of bacterial strains resistant to antibiotics and the difficulty in finding effective therapies against certain viruses have encouraged scientists to seek alternative treatments. Consequently, the potential utility of LF as an antimicrobial agent is very important, and will facilitate the design of new treatments for certain virulent organisms that threaten human safety and health.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Authors’ contributions
EMR, VNU, and EME wrote the draft, tabulated the data, and collected some references; HA collected the literature, organized it, and extracted the main points; EMR designed the work. EMR and VNU constructed the figures, revised the draft, and put together the final version. All the authors read and approved the final manuscript.
Acknowledgments
We extend our appreciation and thanks to all our colleagues who helped and supported us in this work.


