1 Introduction
Cysteine proteases are found widespread in nature and are involved in various biological processes such as processing and degradation of proteins, programmed cell death and response to environmental stimuli [1–4]. The proteolytic activity of cysteine protease is tightly controlled and inhibited by cystatins [5,6]. The inhibitory mechanism of cystatin involves a tripartite wedge formed by Gly residue near the N terminus and two hairpin loops carrying a conserved QxVxG motif and Try residue [7].
Phytocystatins (PhyCys) are plant proteins that inhibit cysteine proteases by direct interaction with an active site. PhyCys was first characterized in rice seed [8]. Moreover, PhyCys play an important role in the regulation of endogenous Cys proteases during plant growth and development, senescence, and programmed cell death [2,9,10]. Several investigations suggested the role of PhyCys in response to cell damages, fungal infection and/or jasmonic acid treatment [11]. In addition, they are also involved in the defense against biotic agents such as viruses [12,13], nematodes, mites [14], and insects [14,15], as well as abiotic stresses including cold, drought, salt, and heat stress [11,16,17]. Numerous studies have underlined the potential role of PhyCys in the development of insect or nematode-resistant transgenic crops [18–20] or as exogenous effectors for the control of cysteine proteases in biotechnological systems of industrial and medical interest. The improved tolerance of cystatin-expressing plants to different abiotic stress cues [21–23] clearly suggests the potential of these proteins to modulate endogenous Cys protease-mediated processes for crop improvement purposes.
In animals, cystatins have been studied intensively and are classified into three families, according to their amino acid sequences. The first stefin family consists of proteins lacking both a signal peptide and disulfide bonds [24]. The second cystatin family is characterized by a signal peptide for secretion and four conserved cysteine residues forming two disulfide bonds. The third one is the kininogens family, which contains three repeats of stefin-type proteins. All these families have the signature QxVxG, which is generally located in the central region of the protein sequence. Two other amino acids are less conserved: a glycine residue located within the N-terminal region and a tryptophan residue within the C-terminal region [17,25,26]. PhyCys share features of the first two families because they have peptide signals but lack disulfide bonds. Generally, their amino acid sequences showed higher similarity with the family II cystatins.
In this study, we have identified a novel full-length sequence of cystatin gene CC14 in maize leaves. In addition, the present work describes the recombinant protein expression, purification and functional characterization of CC14 protein. Transcript abundance for CC14 was analyzed in control and ozone-treated maize leaves by qRT–PCR. Our results indicate for the first time that maize plants adjust CC14 in response to ozone stress and purified recombinant protein CC14 strongly inhibited commercial papain and maize leaf cysteine proteases, thus CC14 could be used for the construction of transgenic plants.
2 Material and methods
2.1 Nucleic acid isolation
In 2008, maize plants (Zea mays L. cv. NK Perform) were grown in a field plot at the experimental farm of AgroParisTech (Thiverval-Grignon, France). Seventy-one days after sowing, the plants were artificially exposed to ozone by means of a linear free-air fumigation device as described previously by Ahmad et al. [3]. Total RNA was extracted from control and ozone-fumigated leaves using the PureLink™ RNA Mini Kit (Invitrogen, France), according to the manufacturer's instructions and protocol described by Ahmad et al. [3,4]. To obtain DNA-free RNA, extracts were treated with the TURBO™ DNase (Ambion, France). RNA quality was assessed by running RNA on 1.5% agarose gel stained with ethidium-bromide, while its concentration was determined with the Nanodrop at A260 (Nanodrop, Noryx, USA).
2.2 cDNA cloning and sequence analysis
cDNA was prepared from 2 μg of total RNA using oligo (dT) primers and reverse-transcriptase using an Omniscript kit from Qiagen company. A cystatin full-length fragment (from first codon ATG to last codon TAA) was PCR-amplified from 50–100 ng of cDNA, using forward primer 5′-GACGACGACAAGATGGCATTCCTCAGCACGAA-3′ and reverse primer 5′-GAGGAGAAGCCCGGTTACAGGAAGTTCAGATAA-3′, (MWG-Biotech, Ebersberg, Germany) and then cloned into pGEM-T Easy vector (Promega, France) and sequenced on both strands (Genoscreen, Lille, France). Nucleotide sequence identity was determined using the online BLASTn program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The presence of signal peptide sequences in the deduced protein sequences was tested using the SignalP 3.0 program (http://www.cbs.dtu.dk/services/SignalP). Multiple-sequence alignments were performed using the ClustalW2 program. The phylogenetic tree for homologous relationship of the selected cystatins was constructed using the ClustalW2 program (http://www.ebi.ac.uk/Tools/phylogeny/clustalw2_phylogeny).
2.3 Cloning and purification of recombinant maize cystatin CC14
The ORF of the full-length cystatin cDNA CC14 (Accession No. JF290478) was cloned into a bacterial expression vector (pET30 EK/LIC, Novogen) and used to transform Escherichia coli cells [strain BL21 (DE3) pLysS, Novogen]. The in-frame insertion of the CC14 cDNA in expression vector was verified through sequencing (Genoscreen, Lille, France). Transformed cells were cultivated in LB at 37 °C and pelleted down at 4000 g for 30 min. The recombinant protein in the supernatant was purified by chromatography on Ni-NTA affinity columns (Qiagen) and was denoted as “CC14” protein. Analysis of the recombinant histidine-tagged CC14 was performed on a PhastSystem, using 8–25% polyacrylamide gradient gels (GE Healthcare, France).
2.4 Recombinant cystatin CC14 blocking activity assay
The enzymatic activity of recombinant CC14 was tested as its inhibitory activity on papain (Roche, France), with azocasein as a substrate. Reaction mixtures (400 μL) containing 500 mm Tris-HCl, pH 6.0, 0.2% (V/V) β-mercaptoethanol, 2.5 μg papain and 2.5 μg recombinant CC14 (absent in control assays) were incubated at 37 °C for 15 min. After addition of 500 μL of a 1% (V/V) azocasein solution, the mixtures were further incubated at 37 °C for 1 h. The reactions were stopped by the addition of 100 μL of TCA (100%) and were transferred into ice for 20 min. After centrifugation at 2000 g for 5 min, the supernatants were collected and measured at A340.
The inhibitory property of recombinant CC14 was tested against the total leaf papain-like cysteine protease (PLCP) activity in maize leaves. The measurement of cysteine protease activity in maize leaves and natural inhibitor CC14 was done according to the protocol described by Ahmad et al. [4]. Briefly, total leaf proteins were extracted in an optimized buffer containing 100 mM of HEPES pH 6.8, 150 mM of NaCl, 0.1% (w/v) of CHAPS, 10% (v/v) of glycerol, 10 mM of DTT and 50 mM of CaCl2, 1 mM of pefaboloc SC, 1 μM of pepstatin A, and 10 mM of EDTA. The cysteine protease activity in the total leaf protein extract was assayed in 200 μL of optimized buffer (100 mM phosphate citrate, pH 6) using synthetic substrate Boc-Val-Leu-Lys-AMC (Bachem, Switzerland). In order to measure the inhibitory activity of CC14, 2 μg of recombinant CC14 were added to the reaction mixture. Substrate fluorescence was checked at λ380 (excitation) and λ460 (emission) by TECAN infinite M1000 (Switzerland). The kinetics of substrate hydrolysis was shown to be linear for 20 min. All assays were carried out in triplicates and no extract controls were used to account for the spontaneous breakdown of the substrate. The inhibition of total leaf cysteine protease by CC14 was calculated as the percentage of inhibition of the total cysteine protease activity.
2.5 Ozone fumigation and CC14 gene expression analysis under ozone stress
Ozone treatments were done to field-grown maize plants according to the protocol explained by Ahmad et al. [3,4]. Briefly, ozone exposure was calculated as the concentration over a threshold of 0.04 ppm (AOT40) cumulated during one or several hour(s) in daylight with a global radiation of 50 W·m2 or greater, as defined for critical levels of ozone vegetation effects in Europe. After 20 days of fumigation, segments from leaves 10 (ear leaves) and leaves 12 (located two rows above the ear) were collected and immediately frozen at −80 °C. AOT40 was 13.1 and 12.8 ppm·h in leaves 10 and 12, respectively. TaqMan PCR was used to detect reference and target gene transcript accumulation, in control and ozone-fumigated leaf samples, using a Light Cycler 2.0 (Roche, France). The primers and TaqMan probes were as follows: for the cystatin gene (CC14)–forward primer: 5′-CGCTGCTGTCACTGGATCA-3′, reverse primer: 5′-CGGCGACCACCTGTTCC-3′, TaqMan probe: YAKCACCACGCTGAGCGGACAGTATGTCABBQ; for the reference gene (S19)–forward primer: 5′-CATTGAAGACTGCGAGGTTC-3′, reverse primer: 5′-CCTTGGGATCGACATCAATGAT-3′, TaqMan probe: FAM: CAAGAGCAGTGGTGCCATTTCACGC–BBQ (TIB MOLBIOL, Berlin, Germany). Ten-μL PCR reaction aliquots containing the cDNA template, obtained from 50 ng of total RNA, 5 pmol of each primer and probe and 2× Taqman Master Mix buffer (Roche, France) were prepared and subjected to the following conditions: 1 cycle at 95 °C for 10 min, 45 cycles at 95 °C for 20 s, 59 °C for 1 min, and 72 °C for 20 s. Cloning of the amplified PCR product was performed using the pGEM-T easy vector plasmid system (Promega, France) following the manufacturer's instructions and used to transform competent cells of Escherichia coli strain GT869 by heat shock. After selection of the recombinant bacteria, plasmid DNA was isolated using Minipreps DNA purification system kits (Promega, France). Sequencing was performed on both strands by Genoscreen (Lille, France). For each cDNA sample, the relative expression level of target genes was normalized by reference to the S19 housekeeping gene and the transcript assay ratio of a target gene to reference gene was determined according to the protocol described previously by Ahmad et al. [3,4]. At least three ozone-treated and control plants (biological repeats) were analyzed using a multifactor analysis of variance (ANOVA) and comparison among means was determined by Tukey's test (P < 0.05). The data were processed using the R statistical analysis system (http://www.r-project.org).
3 Results and discussion
3.1 Isolation and sequence analysis of cDNA encoding a putative cystatin CC14 from maize leaves
In the present study, we report the isolation and characterization of a cystatin cDNA from maize leaves (CC14) and its expression in response to tropospheric ozone stress. The search for the equivalent of CC9 cystatin cDNA (CAG29028) corresponding to a stress-responding cystatin gene in maize [27], using CC9-specific PCR primers, led to the identification of a unique cDNA in maize cv. NK Perform, denoted as CC14 and registered in NCBI with accession number JF290478. Sequence analysis of CC14 showed that it is a full-length cDNA encoding a protein of 177 residues. Cystatins in plants differ from animal cystatins due to the presence of a plant-specific sequence, or PSS, [LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-x-[EDQV]-[HYFQ]-N located in a region corresponding to a predictable amino-terminal α helix [25]. The CC14 protein identified in this study possessed a PSS sequence (Fig. 1). Moreover, phytocystatins can be divided into three subgroups in order to facilitate molecular and evolutionary analysis [28]: group-1 phytocystatins, which contain only 1 cystatin domain with almost 100 residues; group-2 ones, which display 200–250 residues that share a highly conserved cysteine domain at the N terminus with an extended cystatin-like domain at the C terminus; and group-3 ones, which are multicystatins with several repetitive cystatin domains. Most cystatins are composed of only one cystatin domain of about 100 residues in a molecular mass ranging from 12 to 28 kDa [29]. The cystatin CC14 identified in the current study showed similarity to the group-1 phytocystatins due to the fact that it contains only one cystatin domain. However, several cystatins of 85 to 87 kDa with eight cystatin domains have been described in the literature [30]. Each functional cystatin domain has three conserved motifs for inhibition of target enzymes:
- • tripartite wedge containing glycine residue;
- • two hairpin loops carrying a conserved QxVxG motif;
- • tryptophan residue near C-terminal [31].
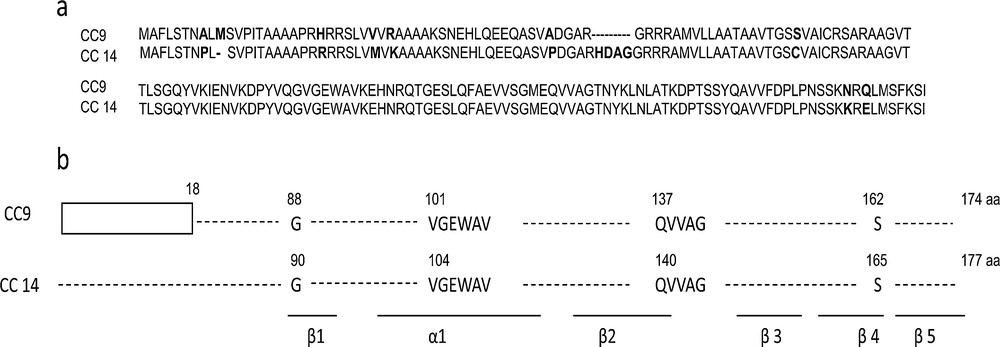
Schematic representations of the deduced CC14 proteins: a: the maize cystatin CC14 deduced amino acid sequence (JF290478, this study) was aligned with that of the maize cystatin CC9 (CAG29028). Amino acids that differ between the two sequences are in bold letters; b: the structure of both deduced proteins were compared. The predicted signal peptide in CC9 is boxed. The approximate location of the five β-sheets and single α-helix is indicated. The cystatin plant-specific sequence LARFAV, the active site QxVxG, and the conserved G residue are also indicated. Masquer
Schematic representations of the deduced CC14 proteins: a: the maize cystatin CC14 deduced amino acid sequence (JF290478, this study) was aligned with that of the maize cystatin CC9 (CAG29028). Amino acids that differ between the two sequences are in bold ... Lire la suite
The CC14 protein identified in this research also contained all three conserved motifs (Fig. 1). The presence of these motifs in the amino acid sequence of the full-length CC14 suggested that the full-length of the gene is probably functionally active in papain and plant cysteine protease inhibition (Table 2). Alignment of CC14 deduced amino acid sequence with the thirteen previously identified maize cystatin sequences showed that CC14 and CC9 were much similar (93%). The next most similar sequence was that of CC12, but even then with only 56% homology. The main differences between CC9 and CC14 remained in the absence of a signal peptide in CC14, and in a 4-amino acid-insertion (HDAG) between CC9 residues R52 and G53 in CC14 (Fig. 1a). The locations of the five β-sheets and single α-helix and of the plant-specific cystatin signature (LARFAV) were fairly identical in both CC9 and CC14 (Fig. 1b). Possible inhibition capacity against papain-like proteases was suggested by the presence of a G residue in the N-terminal part of the molecule and a reactive site QxVxG, located between β2 and β3 sheets. Together, these results suggested that CC14 was an active single-domain cystatin.
In vitro inhibitory activity of CC14 against commercial papain. The recombinant CC14 cystatin was tested on papain in reaction mixtures containing 2 μg of CC14, 2.5 μg of papain and the non-specific substrate azocasein.
| Papain | CC14 | Azocasein | A 340 | % Inhibition |
| − | − | + | 0.367 ± 0.05 | – |
| + | − | + | 0.751 ± 0.13 | – |
| + | + | + | 0.356 ± 0.09 | 100 |
3.2 Homology of the CC14 gene with other cystatins
In order to evaluate the evolutionary relationships, a number of cystatin sequences identified and characterized in other plant species and the newly identified cystatin “CC14” were aligned to construct an unrooted tree. We collected fourteen sequences of maize cystatins CC1 to CC14; thirteen cystatins of barley Hv-CPI-1 to Hv-CPI-13; two cystatins of Arabidopsis AtCYS-4, AtCYS-5; three cystatins of wheat WC1, WC4, WC5; two cystatins of sorghum SbCPI-3, SbCPI-4; one cystatin from Vigna unguiculata and one cystatin of Cakile maritime from the NCBI database to construct a phylogenetic tree (Fig. 2). A phylogenetic tree showed that CC14 (Zea mays L.) has the highest homology with CC9 and CC8. The minimum number of amino acid identities of CC14 (14%) was found with CC4, CC5, and CC11 (Table 1), whereas the maximum number of amino acid identities (94%) was observed between CC14 and CC9 followed by CC14 and CC8 (56%). As shown on Fig. 2, the novel corn cystatin CC14 was in distant parts of the tree compared to barley cystatins. CC14 formed a clade with CC9 and CC12, which, in turn, clustered closely with a maize cystatin involved in defense against cold and drought stress [27]. CC6, CC7 and CC8 were quite isolated in the tree and not closely related to CC14.
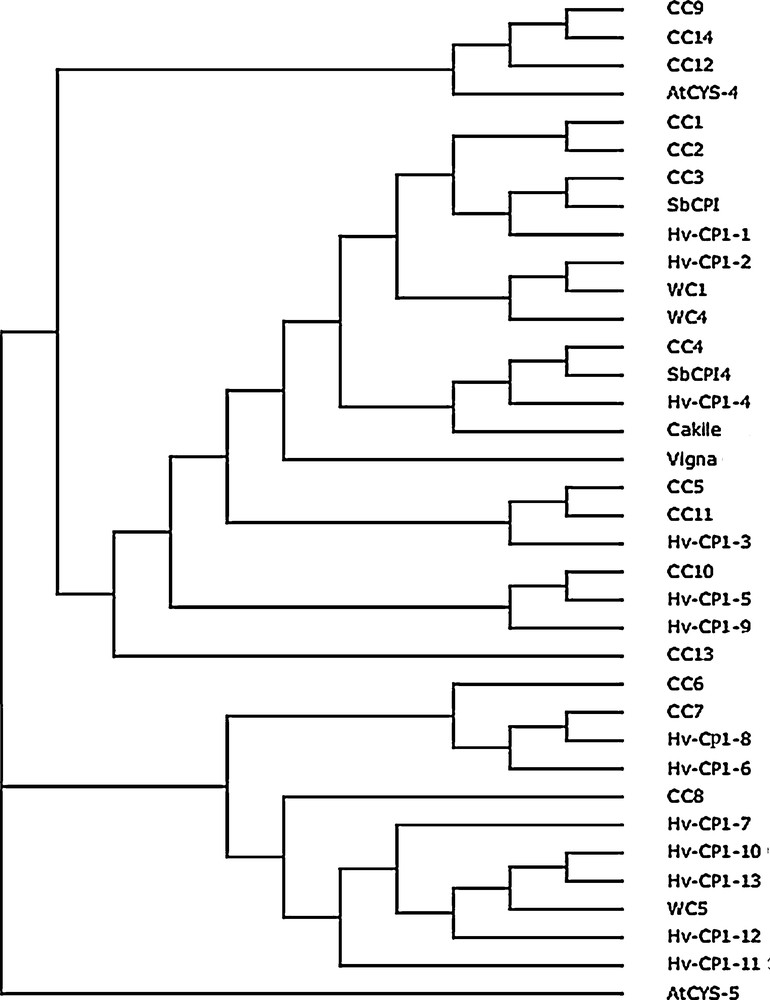
Phylogenetic relationships between several cystatin protein sequences from various plant species, using the Geneious v4.8 program from CLUSTAL W alignments. The neighbor-joining method was used to construct the tree. The cystatin sequences were retrieved from GenBank and included 14 maize (Zea mays L.) sequences CC1 to CC14, (BAA09666, BAA07327, CAG29023, CAG29024, CAG29025, CAG29026, CAG29027, CAG29030, CAG29028, CAG29029, JF290478), 13 barley (Hordeum vulgare L.) sequences Hv-CP1-1 to Hv-CP1-13, (CAA72790, CAG38123, CAG38124, CAG38130, CAG38126, CAG38127, CAG38131, CAG38129, CAG38125, CAG38128.1, CAG38132, CAG38133, CAG38134), two Arabidopsis thaliana sequences (AtCYS-4, Q84WT8; AtCYS-5, Q41916), three wheat (Triticum aestivum L.) sequences (WC1, BAB18766; WC4, BAB18767; WC5, AAL56401), two sorghum (Sorghum bicolor L.) sequences (SbCPI-3, XP_002441309; SbCPI4, XP_002457629), one cowpea (Vigna unguiculata L.) sequence (Vigna, AAQ14319) and one Cakile maritima sequence (Cakile, ABN42208). Masquer
Phylogenetic relationships between several cystatin protein sequences from various plant species, using the Geneious v4.8 program from CLUSTAL W alignments. The neighbor-joining method was used to construct the tree. The cystatin sequences were retrieved from GenBank and included 14 maize ... Lire la suite
Homology of CC14 with other maize cystatins.
| CC1 | CC2 | CC3 | CC4 | CC5 | CC6 | CC7 | CC8 | CC9 | CC10 | CC11 | CC12 | CC13 | CC14 | |
| CC1 | 100 | |||||||||||||
| CC2 | 96 | 100 | ||||||||||||
| CC3 | 53 | 51 | 100 | |||||||||||
| CC4 | 34 | 33 | 41 | 100 | ||||||||||
| CC5 | 36 | 37 | 36 | 28 | 100 | |||||||||
| CC6 | 32 | 31 | 25 | 33 | 29 | 100 | ||||||||
| CC7 | 29 | 29 | 25 | 26 | 26 | 52 | 100 | |||||||
| CC8 | 35 | 34 | 28 | 25 | 21 | 50 | 45 | 100 | ||||||
| CC9 | 24 | 25 | 21 | 16 | 14 | 40 | 29 | 41 | 100 | |||||
| CC10 | 29 | 28 | 30 | 31 | 17 | 30 | 32 | 30 | 20 | 100 | ||||
| CC11 | 25 | 25 | 18 | 16 | 17 | 37 | 27 | 29 | 92 | 22 | 100 | |||
| CC12 | 16 | 16 | 15 | 18 | 12 | 29 | 20 | 22 | 56 | 15 | 13 | 100 | ||
| CC13 | 20 | 20 | 18 | 14 | 12 | 20 | 21 | 14 | 22 | 7 | 20 | 10 | 100 | |
| CC 14 | 18 | 18 | 15 | 14 | 14 | 31 | 25 | 30 | 94 | 18 | 14 | 56 | 21 | 100 |
3.3 Purification and in vitro activity assay of recombinant maize CC14
The recombinant protein, corresponding to cystatin cDNA, was expressed in E. coli cultures and is denoted as “CC14”. It was purified to apparent homogeneity by Ni–NTA affinity chromatography, as shown by the single band detected in the Comassie blue stained polyacrylamide gel (Fig. 3). The apparent Mr of the corresponding His-tag polypeptide was approximately 25 kDa (20 kDa CC14 + 5 kDa His-tag). Several researchers have shown that His-tag did not affect biological or biochemical cystatin activity [32,33]. As shown on Fig. 3, the protein appeared as one primary band of approximately 25 kDa on SDS-PAGE, which closely matched the calculated MW of 25 kDa that was determined after purification of the protein on the Ni2+-affinity resins. The purification of the soluble recombinant protein made it possible to investigate the biochemical activity of CC14. Therefore, activity of recombinant CC14 was assayed in vitro by testing its blocking capacity on the proteolytic activity of papain using azocasein (a chromogenic substrate) and on total cysteine protease activity in control and ozone-fumigated maize leaves using synthetic oligopeptide Boc-Val-Leu-Lys-AMC (fluorogenic substrate). Hydrolysis of azocasein by papain led to an increase in absorbance (A340 nm). In the presence of recombinant CC14, A340 nm values were equivalent to those corresponding to intact azocasein, suggesting the total (100%) inhibition of commercial papain activity (Table 2). A similar phenomenon of inhibition of total cysteine protease activity by recombinant CC14 was observed in maize leaves. After addition of CC14 protein in total maize cysteine proteases, 57–65% and 83–86% inhibition of cysteine protease activity was measured in young leaves # 12 and old leaves # 10, respectively. However, the recombinant CC14 protein did not inhibit 100% cysteine protease activity due to the fact that CC14 non-sensitive cysteine protease may exist in maize leaves.
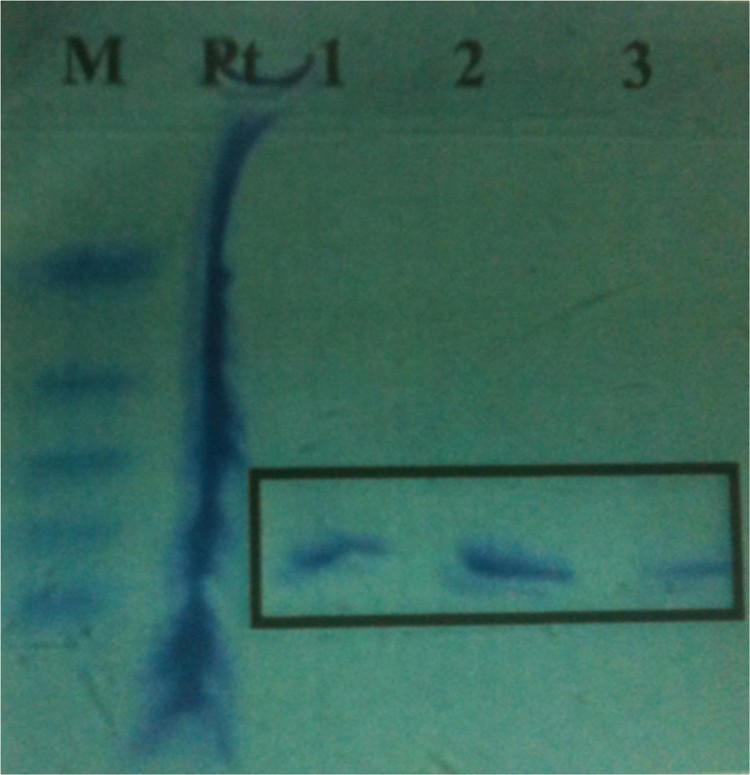
(Color online.) Purification of recombinant maize cystatin (CC14). Molecular marker (M), total protein (Pt) and Ni-NTA-purified CC14 protein of three different extracts (1, 2, 3) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue. The apparent size of the His-tagged recombinant protein was 25 kDa.
3.4 Transcriptional level of CC14 after ozone stress
CC14 gene expression was enhanced three- and four-fold in younger and older leaves, respectively, as compared to control after 20 d of ozone fumigation (Fig. 4). Previous studies have shown that tropospheric ozone induced the expression and enzymatic activity of cysteine proteases involved in the degradation of proteins and programmed cell death in maize leaves [3,4]. In addition, cystatins regulates the activity of endogenous cysteine proteases involved in biotic and abiotic stresses [9,10]. Thus a higher expression of CC14 in maize leaves under ozone stress could have a protective effect on preventing cellular damages induced by excessive cysteine protease activity. Phytocystatins are involved in plant responses to abiotic stresses such as salt [23,34], drought [16] and tropospheric ozone stress (this study). In brief, cystatin mRNA accumulates in the vegetative tissues of barley plants exposed to anaerobiosis, darkness and cold shock [35]; in this study, for the first time, upregulation of cystatin mRNA accumulation was observed under ozone stress (Fig. 4). Cold and heat shocks induced cystatin message accumulation in the leaves and roots of chestnut plantlets [14]. Similarly, in rice seedlings, exposure to the gaseous air pollutant SO2 led to changes in phytocystatin-like proteins. Drought-sensitive and drought-tolerant cowpea plants were submitted to controlled drought stress, dessication, and exogenous ABA [16]. Drought stress induced multycystatin in cowpea (V. unguiculata) leaves and no transcripts were observed in control plants [16]. In maize, 13 cystatin genes (CC1 to CC13) have already been identified that respond differentially to drought and cold stress [36]. The most important drought or cold stress-responsive genes among the 13 identified cystatins were CC8 and CC9 [27] and CC14 identified in this study showed maximum homology with CC9 (Table 1) that could be involved to inhibit cysteine proteases induced by ozone stress.
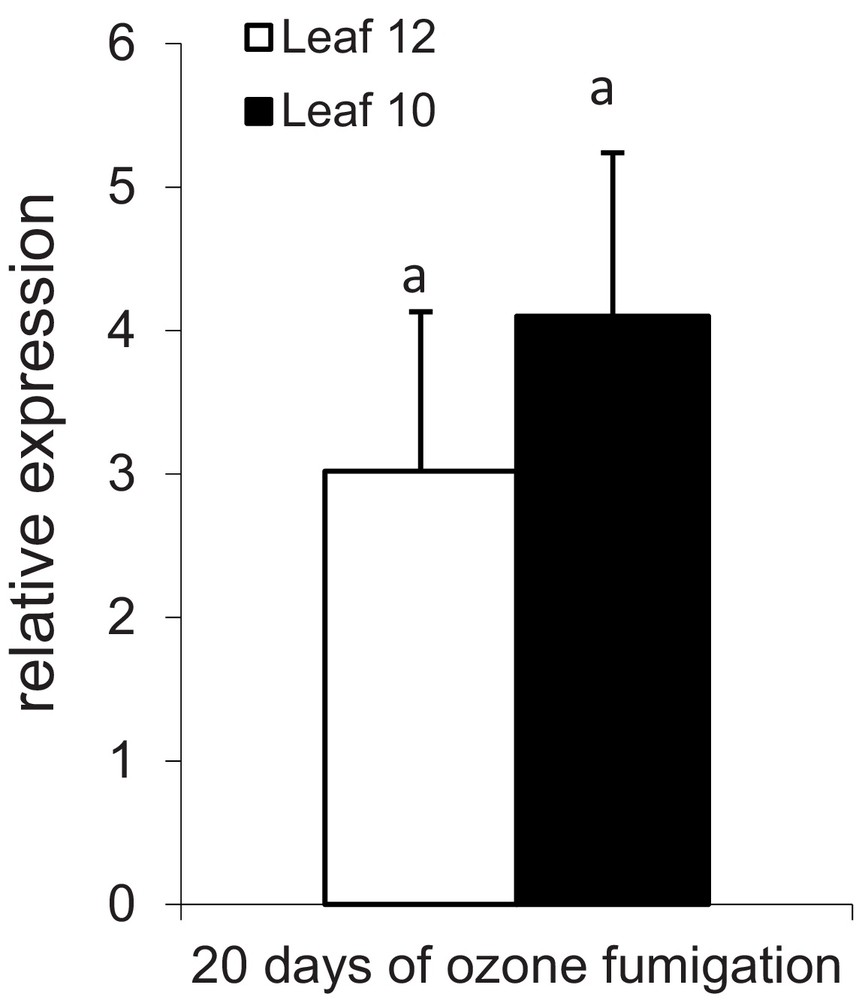
Expression of the CC14 gene in the leaves of field-grown maize plants. Gene expression in leaves of ozone-enriched air, relatively to the corresponding non-fumigated leaves samples (the value of gene expression in non-fumigated leaves is 1). Analysis was performed by TaqMan polymerase chain reaction (PCR), on leaves 12 and 10 collected after 20 days of experiment, using S19 gene as a reference. For ozonated plants, AOT40 was 13.1 and 12.8 ppm·h in leaves 10 and 12 respectively after 20 d of fumigation. Error bars indicate ± SD for three independent replicates. Same letters indicate means that are not significantly different following ANOVA test (P < 0.05). Masquer
Expression of the CC14 gene in the leaves of field-grown maize plants. Gene expression in leaves of ozone-enriched air, relatively to the corresponding non-fumigated leaves samples (the value of gene expression in non-fumigated leaves is 1). Analysis was performed by ... Lire la suite
4 Conclusion
Previous studies have suggested that cystatins are associated with defense, as they play a significant role in protecting host plants from biotic and abiotic stresses [9,10]. In this study, a new cysteine CC14 was identified in maize leaves. Partial characterization showed that CC14 is closely related to drought- and cold-responsive genes (CC9, CC12 and CC8). CC14 gene expression by q RT-PCR increased three- to four-fold under ozone stress. Thus, it could be involved in the regulation of cysteine protease activity in maize leaves. In order to study and verify newly identified cystatin CC14 function in the regulation of cysteine protease activity, we cloned maize CC14 into expression vector pET30 EK/LIC and then expressed in heterologous system E. coli. Purified recombinant protein CC14 was soluble and active and it strongly inhibited commercial papain and maize leaf cysteine proteases involved in the degradation of proteins and programmed cell death. Since, on the one hand, cystatin-expressing plants showed higher tolerance to various biotic and abiotic stresses [23,37,38] and on the other hand, cysteine proteases were shown to be over-expressed in response to ozone stress [3,4,39], thus CC14 inhibitor could be used for the construction of cystatin-expressing plants that could provide higher tolerance to tropospheric ozone [40].
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
This study has been financially supported by the “Agence Nationale de la Recherche”, France through the VULNOZ Project (ANR-08-VULN-012) and the Higher Education Commission (HEC) of Pakistan.


