1 Introduction
In potato tuber, large amounts of proteins, approximately 3 to 6% of the dry weight, are accumulated when they are fully developed and they are relatively stable for several months [1,2]. Excessive amounts of proteins can be accumulated in a discrete deposit (protein body) without any adverse effects on other cellular functions. Potato, therefore, has been extensively studied to accumulate high levels of pharmaceutically or industrially important proteins by using recombinant DNA technology [3,4]. Albumin and globulin constitute most of the soluble proteins in potato tubers [5], and one of the major storage proteins is 11S globulins, known as patatin, dissolving in 0.5 to 1.0 mol of NaCl solution [6]. Patatin constitutes up to 40% of the total soluble proteins in potato tuber, and its molecular weight is about 45 000 kDa [7]. However, one of limiting factors of potato-soluble proteins as nutritional value is their low content of sulphur-containing amino acids, methionine, and cysteine. Patatin contains only 2.0% of methionine and very low cysteine residues out of the total amino acid residues [8,9], which are far less insufficient to meet the animal growth requirement (3.5% of the total supplied protein's weight) recommended by WHO [3,10].
To overcome such a limiting factor, an attempt was reported by Tu et al. [11] for the first time by introducing a gene encoding 2S albumin protein of Brazil nut (BN2S) containing methionine and cysteine contents of 10 and 8 mol%, respectively. When several recombinant gene constructs containing 2 to 7 additional methionine codon created by in vitro mutagenesis were introduced into the genome of potato, and up to 0.2% of the total soluble protein was derived from those introduced constructs in leaves. In tubers, however, much less levels of expression (2- to 4-fold lower levels) were detected. Unfortunately, BN2S is known to be allergenic to human and an alternative heterologous gene encoding methionine-rich protein is therefore necessary to be identified for the development of nutritionally improved potato.
Traditionally, in South Korea, perilla scrap left over oil extraction has been a good supplementary source for animal feeds and plant fertilizer. It has been verified that perilla seeds and leaf contain a rich source of omega-3 polyunsaturated fatty acids (PUFAs), specifically alpha-linolenic acid (ALA) along with omega-6 and omega-9 fatty acids. In addition, perilla oil is the best resource for additional human omega-3 PUFAs [12]. It also contains high contents of various phenolic compounds, resulting in a strong antioxidant activity [13,14].
We have previously characterized a cDNA-encoding 11S legumin-like seed storage protein designated PrLeg from developing perilla (Perilla frutescens), and have identified that it contains relatively high levels (4.24%) of methionine residues based on the nucleotide sequence analysis [15]. This value has been the highest level among 11S legumin-like seed storage proteins reported so far, suggesting that perilla scrap has also been a good source for sulphur-containing essential amino acids to animal. We report here the introduction of the PrLeg cDNA isolated from developing seeds of perilla into potato under a tuber-specific promoter, Patatin. Characterization of transgenic potato indicated the enhancement of methionine content in tubers, suggesting that further studies are necessary to evaluate its nutritional value.
2 Materials and methods
2.1 Plant materials
Potato plant tissues of Solanum tuberosum L. cv. Jowon and cv. Superior grown in a test tube were kindly provided by the National Institute of Highland Agriculture, RDA, South Korea, and maintained in vitro by sub-culturing every four weeks at 23 °C under 16 h light (4000 Lux) and 8 h night conditions [16,17]. The composition of the solid media for sub-culture was 4.4 g/L of MS basic salts [18], 30 g/L of sucrose, and 0.8% of phyto agar. Sterilized leaf explants of 2- to 3-week-old plants were used for plant transformation.
2.2 Vector construction and plant transformation
Previously, a cDNA-encoding PrLeg, an 11S legumin-like storage protein containing high levels of methionine was isolated from the cDNA libraries of developing perilla seeds [15]. The PrLeg cDNA fragment (1610 bp) was originally sub-cloned into the pBK-CMV plasmid. The cDNA fragment was amplified by PCR with pBK-CMV plasmid as a template and primer set (forward, 5′-AAGGTACCAATTCGGCACGAGTCTCTCTCT-3′; reverse, 5′-GGGCTGCAGAAGAAAGAAGTGGATATCTT-3′) designed to include Kpn I and Pst I restriction endonuclease at the terminal end to subclone into a plant vector, pSJ001, kindly provided by Dr Ji-Hoon Ahn from School of Life Sciences and Biotechnology, Korea University, South Korea. pSJ001 vector contains a Bar gene resistant to bialaphos herbicide regulated by the mannopine synthase (Mas) promoter as a selective marker. The Patatin promoter fragment (1539 bp) was amplified by PCR using potato genomic DNA isolated from leaves as a template and a designed primer set (forward, 5′-AAACATTGTTTTATTTTCTCTTTCTTTTT-3′; reverse, 5′-TTTGCAAATGTTCAAAGTGTTTTTAAATTTTGTTGGTGCTTT-3′) from the known nucleotide sequence information (S. tuberosum B 33 gene upstream region, Genbank, accession No. X14483). The amplified fragment was initially cloned into pGEM-T easy vector and subjected as a template for the re-amplification of the final Patatin promoter fragment with another primer set (forward, 5′-GCGCGAATTCATGTTGCCATATAGAGTAGT-3′; reverse, 5′-GCGCGGATCCCATTTTGCAAATGTTCAAAG-3′). The terminal end of this primer set contains EcoRI and BamHI restriction endonuclease site so that the CaMV35S fragment of pSJ001 vector can be substituted with the Patatin promoter.
After the replacement of the CaMV35S fragment to Patatin promoter was confirmed by restriction endonuclease digestion and agarose gel electrophoresis, the PrLeg cDNA fragment was linked to the Patatin promoter at the Bam HI and Xba I restriction endonuclease site. For this purpose, the PrLeg cDNA fragment was re-amplified with a primer set (forward, 5′-GCGGATCCATGGCGTCCAAGCTTCTTCTC-3′; reverse, 5′-GGGTCTAGACTAATAGTGCTTTCTTCCAT-3′) designed to contain the Bam HI and Xba I restriction endonuclease sites at the terminal end. The final recombinant vector was designated as Patatin:PrLeg (Fig. 1). It was transformed into Agrobacterium tumefaciens ASE for potato transformation, as described by [19].
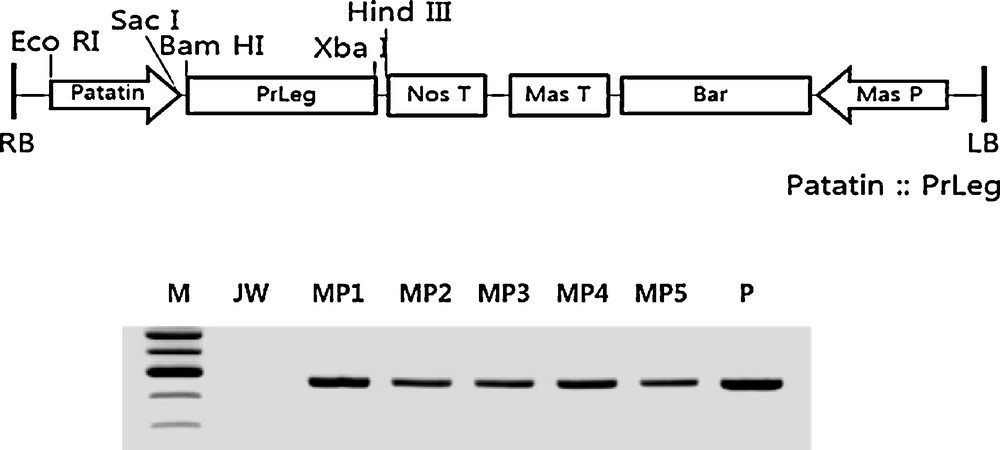
Restriction endonuclease mapping of recombinant T-DNA for PrLeg gene and the result of PCR amplification for PrLeg gene in the studied transgenic plants. Genomic DNA was isolated from a regenerated potato plant on a selective media containing a high concentration of PPT (3.0 mg/L); PCR reaction was conducted with a primer set designed for PrLeg gene and fractionated on a 1.5% agarose gel by electrophoresis. RB, right border; Mas, mannopine synthase; LB, light border; M, size marker; JW, non-transgenic potato cultivar (Solanum tuberosum, cv. Jowon); MP1 to MP5, transgenic lines; P, positive control, recombinant plasmid DNA. Masquer
Restriction endonuclease mapping of recombinant T-DNA for PrLeg gene and the result of PCR amplification for PrLeg gene in the studied transgenic plants. Genomic DNA was isolated from a regenerated potato plant on a selective media containing a ... Lire la suite
The recombinant vector-bearing PrLeg cDNA was then transformed into potato by using leaf explants, as described by Goo et al. [20] and Kim et al. [17]. Briefly, leaf explants were co-cultured with A. tumefaciens carrying recombinant vectors and then transferred to a regeneration medium (MS salts, sucrose 30 g/L, NAA 0.01 mg/L, Zeatin 2.0mg/L, GA3 0.1 mg/L, carbenicillin 500 mg/L). After 8 weeks of regeneration, the induced shoots were transferred to the selection medium containing MS salts plus sucrose 30 g/L, phosphionthricin (PPT) 0.5 mg/L, carbenicillin 250 mg/L, and cultured for a further 4 weeks. To increase the fidelity of transgenic plants, the regenerated plants were re-rooted in a medium containing higher concentrations of PPT (3.0 mg/L) after having cut the root with a razor. The plants were subsequently acclimatized to pots for further experiments, as described by Kim et al. [21].
2.3 Nucleic acid purification, polymerase chain reaction (PCR), Reverse transcriptase PCR (RT–PCR)
For a rapid identification of candidate transgenic plants, genomic DNA was purified from shoots of selective transgenic plants by using the cetyltrimethyl ammonium bromide (CTAB) method [22]. PCR was carried out with purified genomic DNA and primer set for PrLeg gene (forward, 5′-ATGGCGTCCAAGCT-3′; reverse, 5′-CCACCGCATTCGTCAAGG-3′). To detect the level of transcripts, total RNA was purified from the candidate transgenic plants as described by Goo et al. [20]. Three-week-old leaf tissues (0.1 g) or micro-tuber grown in a tissue culture condition for two months (about 1.0–1.5 cm of diameter) was macerated with liquid N2, using a mortar and a pestle. The macerated powder was mixed with Trizol buffer (Invitrogen, Carlsbad, CA) and further purified according to the manufacturer's directions. Five nanogrammes of total RNA were allocated and RT–PCR was carried out with the same primer set of the PrLeg gene by using Suprime script RT_PCR premix provided from Genet Bio. As an internal control, a primer set for actin gene was also designed for forward, 5′-GGCTGGATTTGCTGGTGATG-3′, and for reverse, 5′-CCGCCTGAATAGCAACATAC-3′. Following the fractionation of RT–PCR products by electrophoresis, a photo was taken by using the GelDoc system and the area value of each band was calculated by using the Image J software program (http://rsb.info.nih.gov/ij/index.html). The average of three separate experiments was presented.
2.4 Amino acid analysis
Amino acid analysis was performed at the Feeds and Foods Nutrition Research Center, Pukyong National University, South Korea, using Amino acid analyzer S433 (Sykam, Germany). The apparatus was operated according to the manufacturer's instruction. For sulphur-containing amino acids (methionine and cysteine), the lyophilized powder tuber sample (0.02 g) was treated with 15 mL of performic acid at 0 °C for 24 h and then, 0.75 mL of HBr was added; after the gas had been completely removed, the sample was kept at 0 °C for 1 h. The reactant was then evaporated with a rotary vacuum evaporator. Acid hydrolysis was carried out by adding 15 mL of 6 N HCl, then, the sample was treated with N2 gas for 1 min and incubated at 110 °C for 24 h. The reactant was then filtered through Whatman No. 6 paper and evaporated with the rotary vacuum evaporator. The final sample was dissolved in 25 mL of a sodium citrate loading buffer (pH 2.2) and filtered on a 0.45 μm membrane.
For acid-stable amino acids, 15 mL of 6 N HCl were directly added to an aliquot (0.02 g) of the powered sample in a test tube after lyophilisation; the rest of the procedure was the same as that described for sulphur-containing amino acids. The pre-treated sample was loaded to analyze the amino composition on an Amino acid analyzer equipped with a separation column LCA K06/Na 4.6 × 150 mm S/N 15010303; the operating conditions were: 0.25 mL/min for the reagent flow rate, 0.45 mL/min for the buffer flow rate, and 120 °C for the reactor temperature. Data presented here are the average of the three separate hydrolyses of three individual plants.
3 Results
3.1 Transformation efficiency
We initially confirmed that the recombinant vector, Patatin:PrLeg, was successfully constructed by using the restriction endonucleases digestion pattern (Fig. 1). Then, we transformed it into A. tumefaciens ASE following the transformation of the recombinant T-DNA constructs into the genome of the potato plants. After 4–5 weeks, the young shoots were generated from callus and they were transferred into a root-inducing medium containing 0.5 mg/L of PPT. In order to increase the fidelity of the transformation, the regenerated plants were re-rooted in a medium containing high concentration of PPT (3.0 mg/L) after removing the original root. As a result, we found that the transformation efficiency was very different between the two cultivars. In detail, by using cv. Jowon, 12.5% of the regenerated plants from original explants were able to grow on a strong PPT media (3.0 mg/L), while only 6.7% of the regenerated cv. Superior plants did. However, the results of our PCR analyses showed that 8.3% of the regenerated plants of cv. Jowon produced a positive band for PrLeg gene, but no such a positive band was detected from all the regenerated plants of cv. Superior (Table 1 and Fig. 1).
The efficiency of transformation for PrLeg gene into potato varieties.
| Variety | No. of explant | No. of regenerated plants | No. of resistant plants in PPT (%) | No. of transgenic plants confirmed by PCR (%) |
| Solanum tubersoum L cv. Jowon | 250 | 240 | 30 (12.5) | 20 (8.3) |
| Solanum tubersoum L cv. Superior | 250 | 150 | 10 (6.7) | 0 (0.0) |
3.2 Transcript level of PrLeg gene in the transgenic potato
To confirm the successful integration of PrLeg gene into the genome of the potato and examine the transcript levels of PrLeg gene, five individual plants were randomly chosen and RT–PCR analyses were carried out with specific primer sets designed for PrLeg and an internal control, actin gene. As expected, in the tuber tissue of all the transgenic plants, high levels of PrLeg transcript were detected, but very low level or only a trace level was present in the leaf tissues, while PrLeg transcript was not detected at all in the control non-transgenic plant. In both control and transgenic plants, similar levels of the actin transcript were detected, indicating that the same amounts of total RNA (5 ng) were allocated for RT–PCR analyses. Interestingly, the MP4 transgenic plant accumulated more than a 2-fold higher amount of PrLeg transcript compared to other transgenic plants (Fig. 2).
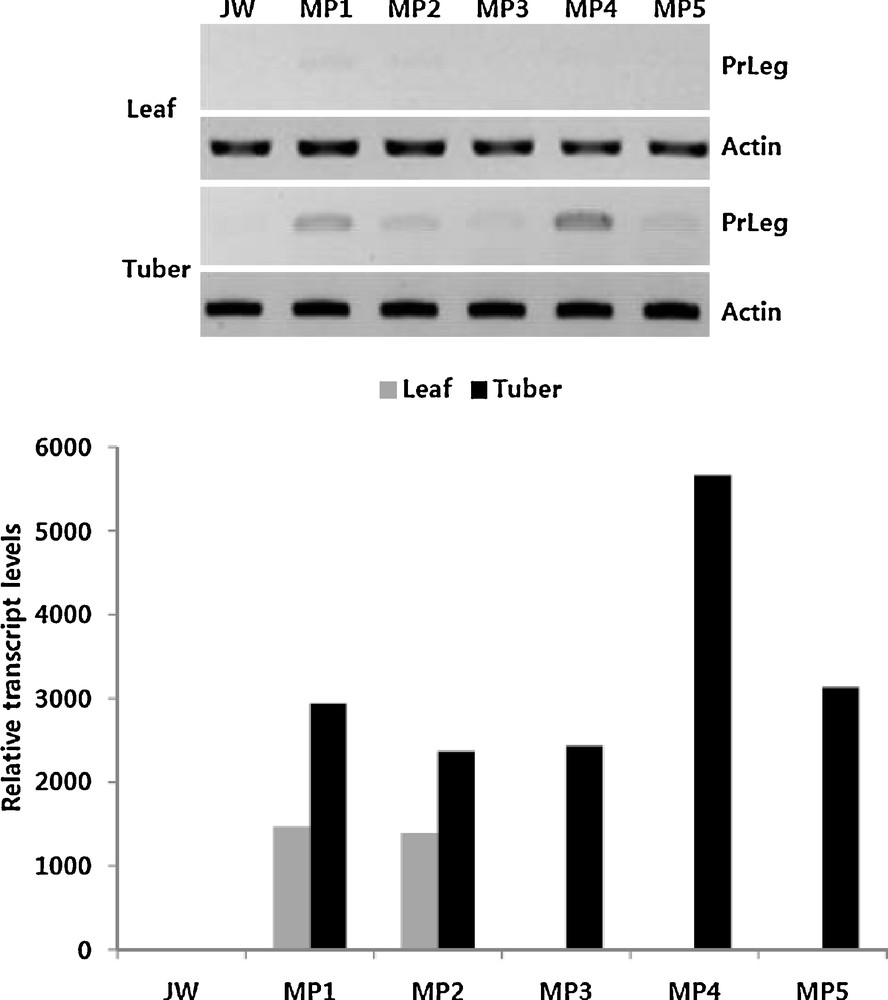
The result of RT–PCR with transgenic potato lines. Total RNA (5 ng) isolated from transgenic potato lines (MP1 to MP5) and a parental line (JW, non-transgenic cultivar) was used to allow RT–PCR and fractionated on 1.5% agarose gel by electrophoresis (upper panel). In the lower panel, relative transcript levels were indicated after calculation of the area value of each band by using Image J software program. Each value is the mean of the three replicate experiments.
3.3 The enhanced level of methionine content in transgenic potato
We then tried to measure the methionine content in micro-tubers induced from transgenic potato plants in a test tube for two months, and we found that most individual transgenic lines showed higher contents of methionine, except the MP4 line when compared with that of the control plant, the non-transgenic parental cultivar. In particular, MP2 and MP5 lines accumulated more than a 3-fold more elevated level of methionine compared to that of the control line (105.4 mg/100 g) (Fig. 3). For other non-sulphur-containing amino acids that were measured without pre-treatment with performic acid and HBr before acid hydrolysis, very similar levels were maintained when compared with those of the control, non-transgenic parental line, except those of asparagine and glutamine that were slightly enhanced (Table 2).
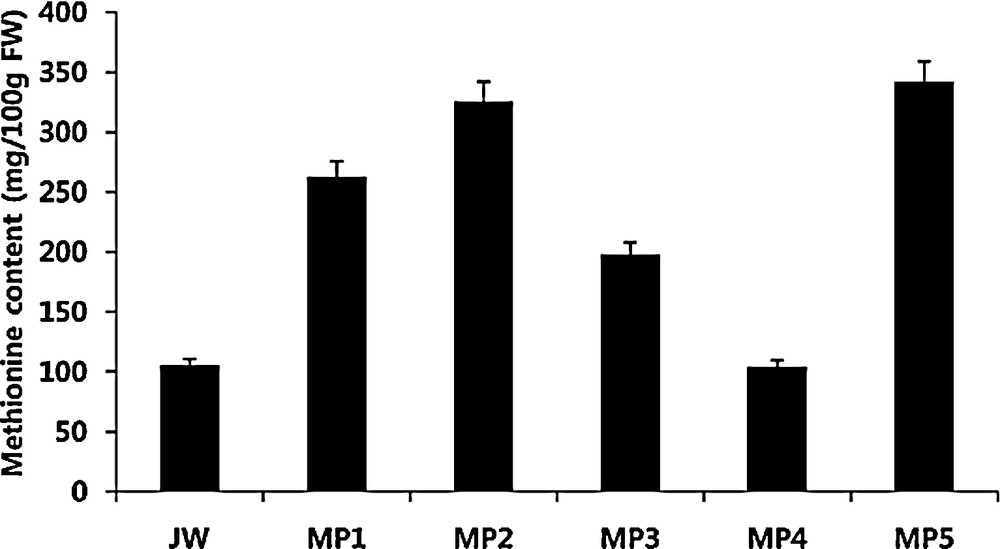
Methionine content of transgenic potato lines. The lyophilized powder tuber from each line (Mp1 to MP5) and non-transgenic line (JW) were treated with performic acid following acid hydrolysis, and the amount was measured with the amino acid analyzer as described in section Materials and methods. Each value is the mean of the three replicate experiments.
Amino acid composition (%mol/mol) of potato tuber proteins. Data presented are the average of three separate hydrolyses of three individual plants.
| Asp | Thr | Ser | Glu | Pro | Gly | Ala | Val | Ile | Leu | Tyr | Phe | His | Lys | Arg | |
| JW | 0.30 (0.10) |
0.05 (0.01) |
0.06 (0.00) |
0.23 (0.05) |
0.03 (0.00) |
0.05 (0.00) |
0.05 (0.00) |
0.07 (0.00) |
0.05 (0.00) |
0.09 (0.00) |
0.02 (0.00) | 0.07 (0.00) |
0.07 (0.00) |
0.08 (0.00) |
0.13 (0.00) |
| MP5 | 0.40 (0.08) |
0.04 (0.01) |
0.05 (0.01) |
0.31 (0.09) |
0.05 (0.00) |
0.04 (0.00) |
0.05 (0.00) |
0.08 (0.01) |
0.06 (0.01) |
0.07 (0.00) |
0.06 (0.01) |
0.05 (0.01) |
0.04 (0.01) |
0.09 (0.02) |
0.07 (0.01) |
4 Discussion
In this study, we tried to over-express a novel gene, PrLeg encoding legumin protein cloned from perilla under the direction of a tuber-specific promoter, patatin, to increase the content of a sulphur-containing essential amino acid, methionine. The percentage of sulphur-containing essential amino acids in PrLeg polypeptide is relatively high (5.3%) based on the total residue number of each amino acid predicted from the nucleotide sequence of cDNA and their respective molecular weights [15]. For instance, the legumin storage proteins from other crops, such as rice, soybean, amaranth, and potato exhibit limited values for sulphur-containing amino acids, only 1.8–2.2%, which are much less to meet the values recommended by WHO (Table 3). In addition, PrLeg protein contains high levels of isoleucine (3.8%) and valine (6.4%) compared to those of patatin protein, which is a major storage protein in potato tuber. Patatin protein contains only 2.6 to 2.7% of isoleucine and valine, which are far less to meet the requirement of WHO, but the content of lysine is relatively high enough compared to rice, soybean, and rapeseed. Therefore, in this study, PrLeg has been considered as an ideal polypeptide for the enhancement of the nutritional value by over-expression in potato tubers (Table 3).
Percentage of essential amino acids in PrLeg, AmA1, and patatin, in comparison with the values recommended by the World Health Organization.
| PrLeg (Perilla) |
Patatin (Potato) |
AmA1 (Amaranthus) |
WHO | |
| Trp | 0.8 | 0.6 | 3.6 | 1.0 |
| Met/Cys | 5.3 | 2.2 | 3.9 | 3.5 |
| Thr | 3.2 | 3.8 | 5.1 | 4.0 |
| Ile | 3.8 | 2.6 | 6.1 | 4.0 |
| Val | 6.4 | 2.7 | 5.2 | 5.0 |
| Lys | 3.0 | 4.2 | 7.5 | 5.5 |
| Phe/Tyr | 6.4 | 7.1 | 13.7 | 6.0 |
| Leu | 7.5 | 6.2 | 9.2 | 7.0 |
| Reference | Jin et al., 2000 [15] | Racusen and Foote, 1980 [41] | Raina and Datta, 1992 [42] | Senft, 1980 [43] |
As we expected, it was found that most of transgenic plants accumulated the higher level of PrLeg transcript in their micro-tuber under the tuber-specific promoter, while only a trace or very little amounts were detected in their leaf tissues. However, the increased levels of methionine in individual transgenic plants were not exactly correlated with the accumulated level of their transcripts of introduced PrLeg gene in tubers. In detail, MP4 transgenic line showed the highest levels of transcript, while the methionine accumulated amount was the lowest, almost the same level as for a control, non-transgenic plant (Figs. 2 and 3). For the moment, we have no clear answer to this observation, but it is assumed that the active transcription of the introduced gene in transgenic plants may cause the inhibition of the following processes. Nevertheless, individual transgenic plants (MP1, 2 and 4) might be good candidates for further examination since they accumulated 2.5 to 3.5-fold more elevated amounts of methionine compared to non-transgenic potato (JW).
Previously, a number of genes encoding methionine-rich proteins have been introduced into several crops for the enhancement of their nutritional value in either vegetative tissue or storage organ of potato [11], legumes [23], canola [24,25], lupine [26], and forage crops [27,28]. For example, Zein, a seed storage protein of maize that is known to contain the highest methionine content up to 28% of the total amino acid residues (37 g 100 g−1 of protein) [29] and 2S albumin protein of Brazil nut (BN2S), and which is also known to be a methionine-rich protein containing up to 18% (23 g 100 g−1 of protein) of the total amino acid residues [30] have been over-expressed in several plants, and 2- to 6-fold enhanced levels of methionine contents have been reported. However, no successful achievement has been reported with potato tuber until Chakraborty et al. [31] tried to over-express a gene encoding seed albumin of Amaranthus hypochondriacus (AmA1) into potato plants under the direction of granule bound starch synthase (GBSS) promoter. They were able to increase the level of methionine content up to 3- to 7-fold in potato tuber tissue compared to the non-transgenic parental line. Interestingly enough, the percentage of sulphur-containing amino acids (methionine and cysteine) in AmA1 polypeptide was only 3.9%, which is less than that of PrLeg (5.6%) and much less than those of BN2S or Zein (28 and 18%, respectively). In addition, AmA1 polypeptide contains higher levels of of Thr. Ile. Lys, Phe, Tyr, and Leu compared to those of PrLeg (Table 3) and led to increase the levels of those amino acids in transgenic potato tubers when AmA1 gene was over-expressed [31]. However, we were not able to observe the elevation of other amino acids, while only a slight increase in asparagine and glutamine contents (1.6- to 1.8-fold) was identified. Our observation is not surprising, since the percentages of those amino acids are not high enough in PrLeg protein. Moreover, the content of Lys in patatin protein, a major storage protein in potato, is already sufficient according to the value recommended by WHO (Tables 2 and 3).
One of the major limiting factors for the enhancement of the content of sulphur-containing amino acids in storage organs or tissues of plants is the availability of the free amino acid form in the cytoplasm for incorporation into the polypeptide. It has also been reported that the over-expression of various methionine-rich proteins in vegetative tissues do not lead to a sufficient elevation in methionine levels to meet the requirements for animal feeding. Apparently, the very low natural pool of free methionine in plants may limit the accumulation of methionine-rich proteins [32,33]. Indeed, when such methionine-rich proteins were expressed in seeds, their accumulation was at the expense of other sulphur compounds or of other endogenous methionine-rich proteins [34–38]. In addition, the over-accumulation of BN2S, Zein or sunflower seed albumin (SSA) in target plants may cause another unbalanced amino acid composition since they contain high levels of methionine, but not of the other essential amino acids. This was more evident when the methionine content was dramatically increased (up to 30- to 239-fold) by the manipulation of genes involved in the biosynthetic pathway of methionine [39]. Co-transformation strategy has been applied to accumulate both free methionine and Zein protein, but all the transgenic lines have shown various abnormal phenotypes including severe growth retardation, changes in the leaf architecture and reduction in tuber yield [40].
In summary, our preliminary results clearly indicate that PrLeg gene isolated from perilla can be a good candidate for the enhancement of sulphur-containing essential amino acids in potato tuber and that further nutritional evaluation study along with field tests for their agronomic traits will be worthwhile.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgement
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ008097), Rural Development Administration, Republic of Korea and Year 2013 Grant from Gyeongnam National University of Science and Technology.


