1 Introduction
Ralstonia solanacearum is an important bacterial pathogen distributed worldwide and caused wilt of economically important crop plants, mostly solanaceous in tropical and subtropical as well as temperate regions of the world [1,2]. The soil-borne pathogens colonize in xylem, causing bacterial wilt in a very wide range of potential crop plants [3]. The pathogen has an extended host range including hundreds of plant species in 50 families [4]. R. solanacearum can overwinter in plant debris or diseased plants, wild hosts, seeds or vegetative propagative organs like tubers [5]. The bacteria can survive for a long time in water (up to 40 years at 20–25 °C in pure water) and the bacterial population is reduced in extreme conditions (temperature, pH, salts, etc.). It can also survive in cool weather and enter a state in which they are viable but not culturable [6]. R. solanacearum was classified into five races on the basis of differences in host range [6,7,8], and six biovars according to their hexose oxidase ability versus three alcohols and three disaccharides [2,7,9,10]. Unlike other phytopathogenic bacteria, race systems of R. solanacearum are not based on gene-for-gene interactions. Instead, these are determined based on the pathogenicity of each isolate in different kinds of host plants.
Although the identification based on biochemical tests proved useful and is still accepted as a standard, it is laborious and time-consuming [2,10]. To improve the understanding of the genetic relatedness among R. solanacearum species complexes, DNA-based analysis has been used in recent years. Molecular techniques, such as restriction fragment length polymorphisms (RFLP), amplified fragment length polymorphisms (AFLP) [12,13], and also random amplified polymorphic DNA (RAPD) [4,14,15] have been applied extensively for genotype diversity analysis in R. solanacearum strains. RAPD markers can be detected by the random amplification of genomic DNA fragments of different sizes through the polymerase chain reaction [16]. RAPD technique is easy and reliable and require small amount of DNA (5–20 ng), single short primers of arbitrary sequence, the rapidity to screen for polymorphisms, the efficiency to generate a large number of markers for genomic mapping and the potential automation of the technique. In addition, no previous knowledge of sequence is required [17] and it is a relatively simple and inexpensive method for examining variations in the total genome [18]. Though RAPD has some limitations, it is being used as one of the most powerful techniques for genetic studies, for example, analysis of genetic variation in plants, fungi and bacteria [19], and construction of the first linkage maps for certain species and pathogens [3]. RAPD methods have been successfully used for genetic variability analysis [4,14,15].
Vegetable growers from different regions of Bangladesh has been communicating with Plant Disease Clinic, Department of Plant Pathology, Bangladesh Agricultural University, Mymensingh for diagnosis of wilt problems in tomato, potato, brinjal, and some other vegetable crops and seek advices for a low-cost, effective and eco-friendly management strategy. The main control strategy has been the use of resistant varieties. However, the stability of bacterial wilt resistance in brinjal, potato and tomato is highly affected by pathogen density, pathogen strains and several soil factors. Different research organizations in Bangladesh and AVRDC have been working on the development of bacterial wilt-resistant varieties, but there are still only a few varieties showing stable resistance. The use of bacterial wilt-susceptible commercial cultivars grafted onto bacterial wilt-resistant rootstocks has been demonstrated to be an effective management tool in both the Philippines and Bangladesh [20,21]. However, grafting is not getting popularity at the farmer's level because of the high cost of grafted seedlings, and no other yield benefits over non-grafted seedlings, as shown by several studies conducted by AVRDC. Other control methods such as soil amendments, crop rotation, biological control, and field sanitation are often not effective. Because no single control method will provide good and sustainable control of the disease, integrating different methods to manage the disease is mandatory. A recent biochemical characterization of R. solanacearum isolates causing wilt in brinjal and potato in Bangladesh were reported by our group [22]. Although the ability to predict the biological, ecological, and epidemiological properties of R. solanacearum strains using a meaningful taxonomic classification system could aid in the development of successful management practices, it is clear that a better understanding of the population structure of this highly diverse pathogen is needed in order to develop resistance/tolerant eggplant and potato cultivars through the screening program. In spite of the necessity of the correct assessment of genetic diversity of the field population of this bacterium to design an effective diagnostic protocol and to deploy resistant cultivars, however, very little is known about the extent of genotypic diversity that may exist in a localized population of this bacterium in the country. RAPD markers were used because they have the potential to detect polymorphism in the entire genome as compared to other techniques [23]. RAPD markers have been used for strain differentiation [24], identification and other genetic analysis for several bacterial genera during the last two decades [23,25–28]. Taken together, the present study was undertaken to assess the potentiality of RAPD as a marker for analysing the diversity of R. solanacearum collected from some fields of some selected locations.
2 Materials and methods
2.1 Survey and sample collection
A survey was carried out to know the status of bacterial wilt in Bangladesh in terms of its incidence and severity as well as to collect the isolates of the bacterial wilt pathogen, R. solanacearum. Seven major growing areas, viz. Panchagarh, Rangpur, Pabna, Jessore, Jhinaidah, Jamalpur and Mymensingh, for eggplant, and eight major growing areas, viz. Jamalpur, Munshigonj, Panchagarh, Bogra, Jessore, Chandpur and Nilphamari, for potato, were selected for the survey. At least three locations in each area and five farmers’ fields for each growing area were surveyed to record the bacterial wilt incidence and severity. For a quick field diagnosis of brinjal wilt caused by bacterium R. solanacearum and to distinguish bacterial wilt from vascular wilts caused by fungal pathogen and nematode, bacterial streaming from infected plant material was performed and confirmed by streaming of milky white masses of bacterial cells (ooze). At least, 10 samples of the diseased plants were collected from each of the surveyed area and were brought into the laboratory for the isolation of different groups of isolate of R. solanacearum.
2.2 Assessment of disease incidence and severity
The status of the bacterial wilt of eggplant was assayed based on wilt incidence and severity. Data on wilt incidence were recorded in at least three locations from five farmer fields for each growing area. Then, the percent wilt incidence was calculated using the following formula:
Five plants were randomly selected from each farmer field in each location to calculate wilt severity in each growing area. The severity of bacterial wilt was recorded based on the severity scale as described previously by Horita and Tsuchiya [29]. Briefly, 1 = no symptom, 2 = top young leaves wilted, 3 = two leaves wilted, 4 = four or more leaves wilted and 5 = plant death.
2.3 Isolation, identification and purification of R. solanacearum
The isolation, identification and purification of R. solanacearum were done as described previously by Rahman et al. [22] and Ahmed et al. [1]. Briefly, bacterial oozes coming out from the cut end of each sample were streaked on Nutrient Agar (NA). The plates were then incubated at 28 °C for at least 24 h. After isolation, R. solanacearum isolates were purified by streaking a single colony of each isolate on Triphenyl tetrazolium chloride (TTC or TZC) medium as described by [18]. To confirm the isolates of R. solanacearum, the pathogenicity test was performed in one-month-old seedlings of the respective hosts by the soil inoculation method. A single colony of R. solanacearum showing virulent, fluidal, irregular and creamy white with pink at the centre was selected for each group of isolates for the pathogenicity test. The bacterial suspension (approximately 108 CFU/mL) of each isolate representing a group was injected in the leaves of tobacco plants aged 30–40 days. Bacterial suspensions were injected into the intracellular space of the leaf with a hypodermal syringe. A hypersensitive reaction (HR) was observed daily and continued up to 5 days of the infiltration. The isolates of R. solanacearum were preserved in 10% skim milk kept at −20 °C refrigerator for subsequent biochemical studies.
2.4 Biochemical characterization of R. solanacearum
The virulent (colonies with pink or light red colour or characteristic red centre and whitish margin) and avirulent (smaller, off-white and non-fluidal colonies) strains of R. solanacearum were identified in a triphenyl tetrazolium chloride (TTC) medium containing 0.005% TTC [29]. Several biochemical tests, viz. Gram staining reaction, potassium hydroxide solubility test, Kovac's oxidase test, Levan test, and Sugar fermentation test, were performed for confirmation of the R. solanacearum isolates. A single isolate of R. solanacearum from each group was randomly selected for biochemical tests.
2.5 Determination of biovars and races of R. solanacearum
The seven isolate groups of R. solanacearum were differentiated into biovars based on their ability to utilize disaccharides (sucrose, lactose, and maltose) and sugar alcohols (manitol, sorbitol and dulcitol) as described previously by Hayward [2] and He et al. [8]. The biovars were determined in a mineral medium (NH4H2PO4 1.0 g, KCl 0.2 g, MgSO4·7H2O 0.2 g, Difco Bacto Peptone 1.0 g, agar 3.0 g and bromothymol blue 80.0 mg per litre) containing 1% sugar. About 200 μL of the melted medium is dispensed into the wells of a microtitre plate. Inocula for each group of isolates were prepared by adding several loopfuls of bacteria from 24-h-old cultures to distilled water to make a suspension containing about 108 CFU/mL. Then, 20 μL of the bacterial suspension was added to the wells of the microtitre plate incubated at 28 °C. The tubes were then examined at 3 days after inoculation for change in pH by a colour change [30].
The races of R. solanacearum isolate groups were identified by pathogenicity test on wide host range [31]. Seedlings of tomato and chilli were raised in tray and 1-month-old seedlings (tomato and chili) were inoculated by soil inoculation method. The incubated plants were then kept in the net house until symptoms development. Seven isolate groups of R. solanacearum were collected from wilted brinjal and potato plants from different locations in Bangladesh (Table 1). For identification of the bacterial isolates of R. solanacearum, a series of biochemical tests such as Gram's stain, potassium hydroxide solubility test, Kovac's oxidase test and Levan's test were performed. The isolates were purified on a tetrazolium chloride (TTC) medium according to Kelman [32] and preserved at −20 °C in a refrigerator in 10% skim milk for subsequent studies [27]. Finally, three isolates groups, viz. RsB-1, RsB-2 and RsB-3 from eggplant and four isolate groups, viz. RsP-1, RsP-2, RsP-3 and RsP-4 from potato plants were selected for the analyses of R. solanacearum genetic diversity. The experiment was conducted at the Biotechnology Laboratory of Bangladesh Institute of Nuclear Agriculture, Mymensingh, Bangladesh.
Status of bacterial wilt of eggplant and potato in terms of its incidence and severity in some selected growing areas in Bangladesh.
| Areas Surveyed | Incidence (%) | Severity (1–5 scale) |
Areas surveyed | Incidence (%) | Severity (1–5 scale) |
| Eggplant | Potato | ||||
| Panchagarh | 20.00a | 3.00 | Munshigonj | 22.65a | 3.80 |
| Rangpur | 22.52a | 3.26 | Nilphamari | 19.98b | 3.00 |
| Jessore | 20.56a | 2.93 | Jamalpur | 9.07c | 2.90 |
| Mymensingh | 14.36b | 3.13 | – | – | – |
| LSD | 2.927 | LSD | 1.371 | ||
| Level of significance | a | Level of significance | b |
a Significance at the 5% level of probability.
b Severity data recorded at the time of survey.
2.6 Genomic DNA extraction and quantification
The extraction of genomic DNA of each isolate of R. solanacearum was done according to Chen and Kuo [29]. The concentration of DNA in the extracted samples was quantified through absorbance reading using a UV Spectrophotometer at 260 nm. The quality of the DNA was verified by electrophoreses on a 0.8% agarose gel in TBE (tris–boric acid–EDTA) buffer.
2.7 PCR amplification and electrophoresis
RAPD reactions were performed following the process of Williams et al. [16] with some modifications [16]. The screening of primer was done with 15 arbitrary decamer primers (Bengalore Genei, India). Four primers producing good scorable and reproducible bands were selected for subsequent RAPD analysis of bacterial isolates (Table 2). PCR reactions were done in a 10.0-μL reaction mixture containing 1 X PCR buffer (10 mM of tris HCl, pH 8.5; 50 mM of KCl and 15 mM of MgCl2), 10 mM each of dNTP (Bengalore Genei, India), 5 pmol of primer, 2 U of Taq DNA polymerase (Bengalore Genei), 100 ng of genomic DNA. DNA amplification was carried out in a DNA thermocycler (Biometra, Germany) at the following thermal profile: initial denaturation for 3 min at 94 °C, followed by 41 denaturation cycles of 1 min at 94 °C, 1-min annealing at 37 °C and extension at 72 °C for 2 min. A final extension step at 72 °C for 10 min was allowed for complete extension of all amplified fragments. Amplified fragments were separated on a 1.5% agarose (Invitrogen, Canada) gel in 1 X TBE buffer along with 20 bp DNA weight marker (Bengalore Genei, India) for 90 min at 100 V. The gel was stained with ethidium bromide (Fisher Scientific) solution (0.1 μg·mL−1) for 15 min in a shaker. Finally, the gel was visualized under UV-transilluminator and photographed by Gel Documentation System (Biometra, Germany).
Biochemical tests of Ralstonia solanacearum isolates groups collected from different growing areas of eggplant and potato.
| Isolate Name |
Pathogenecity test and color test on TTC media | HR Test | Gram staining reaction | KOH solubility test | Kovac's oxidase test | Levan test | Sugar fermentation test | Inference | |||
| Dextrose | Sucrose | Manitol | Lactose | ||||||||
| Eggplant | |||||||||||
| RsB-1 | + | + | + | + | + | + | + | + | + | + | Ralstonia solanacearum |
| RsB-2 | + | + | + | + | + | + | + | + | + | + | Ralstonia solanacearum |
| RsB-3 | + | + | + | + | + | + | + | + | + | + | Ralstonia solanacearum |
| Potato | |||||||||||
| RsP-1 | Ralstonia solanacearum | ||||||||||
| RsP-2 | + | + | + | + | + | + | + | + | + | + | Ralstonia solanacearum |
| RsP-3 | + | + | + | + | + | + | + | + | + | + | Ralstonia solanacearum |
| RsP-4 | + | + | + | + | + | + | + | + | + | + | Ralstonia solanacearum |
2.8 Scoring and data analysis
The dominance of RAPD markers indicates that each band represented the phenotype at a single allelic locus [33]. The amplified bands were visually scored as present (1) and absent (0) separately for each individual and each primer. The scores obtained were pooled to create a single data matrix. The calculated data were used to estimate polymorphic loci, Nei [34] genetic diversity, genetic distance and a UPGMA (Unweighted Pair Group Method with Arithmetic Means) dendrogram using computer-based program, POPGENE (Version 1.31) [34,35].
3 Results
3.1 Variations in wilt incidence and severity in some selected growing areas
A total of seven major eggplant growing areas, viz. Panchagarh, Rangpur, Pabna, Jessore, Jhinaidah, Jamalpur and Mymensingh, were surveyed to know the status of bacterial wilt of eggplant in terms of its incidence and severity. However, bacterial wilt infection was noticed in four growing areas at the time of our survey, viz. Panchagarh, Rangpur, Jessore, and Mymensingh .A significant variation was observed in terms of bacterial wilt incidence among the major growing areas surveyed (Table 1 and Fig. 1). The highest (22.52%) bacterial wilt incidence was recorded in Rangpur, followed by Jessore and Pachagarh, with 20.56 and 20.0% wilt incidence, while the lowest (6.12%) bacterial wilt incidence was recorded in Jamalpur. An incidence of brinjal bacterial wilt of 14.36% was recorded in Mymensingh. On the contrary, the highest (3.26) bacterial wilt severity was recorded in Rangpur, while the lowest (2.93) bacterial wilt severity was recorded in Jessore, followed by Panchagarh and Mymensingh, with values 3.00 and 3.13, respectively (Table 1). A total of seven selected potato-growing districts, viz. Jamalpur, Munshigonj, Panchagarh, Bogra, Jessore, Chandpur and Nilphamari, were surveyed to know the status of bacterial wilt of potato in terms of its incidence and severity. However, bacterial wilt infection was noticed in three districts, namely Munshigonj, Nilphamari, and Jamalpur (Fig. 1). A significant variation was observed in terms of bacterial wilt incidence among the selected growing areas surveyed (Table 1). The survey results showed that the highest (22.65%) bacterial wilt incidence was recorded in Munshigonj, followed by Nilphamari (19.98%), while the lowest (9.07%) bacterial wilt incidence was recorded in Jamalpur (Table 1). The severity scale was calculated for each location using five randomly selected plants from each farmer field. The highest (3.80) bacterial wilt severity was recorded in Munshigonj, while the lowest (2.90) bacterial wilt severity was recorded in Jamalpur during the time of survey.

(Colour online) Distribution of R. solanacearum isolates analyzed in this study. Black and red circles indicate the survey areas where bacterial wilt infections were noticed for eggplant and potato, respectively.
3.2 Isolation, identification and confirmation of R. solanacearum isolates
A total of 51 R. solanacearum isolates were obtained from the wilted brinjal plant samples collected from the different locations surveyed. Twenty-four isolates were obtained from Panchagarh, 15 from Rangpur, eight from Jessore and four from Mymensingh. A total of 44 R. solanacearum isolates were obtained from the wilted potato plant samples, i.e. 20 from Munshigonj, 17 from Nilphamari, and 7 from Jamalpur. Although equal numbers of infected plants were collected from each one of the surveyed areas, the number of isolates varied because of the failure of isolation of the bacterium from all the infected plants or because of the growers’ management practices that may kill the bacterium. All of the R. solanacearum isolates collected from wilted eggplant and potato plants produced cream-colour colonies on NA media after 24 h of inoculation. Biochemical tests revealed that all isolate groups were R. solanacearum (Table 2). The results of pathogenicity tests revealed that all the isolate groups of R. solanacearum from eggplant were able to cause wilt symptoms in eggplant seedlings and those obtained from potato only able to cause wilt symptoms on potato plants (Table 2). The isolates of R. solanacearum collected from the wilted brinjal plants were tested for hypersensitive reaction in tobacco. The results showed that all the isolates were able to cause the rapid death of local tissue cells between veins of tobacco leaves (Table 2).
3.3 Differentiation of R. solanacearum isolates into biovars and races
To assess the phenotypes of R. solanacearum, isolates collected from field, biovars and races of the isolates were determined. The biovar of all seven groups of R. solanacearum isolates obtained from both eggplant and tomato were identified by utilization of disaccharides and of hexose alcohols. The result of the biovar test showed that all seven groups of R. solanacearum isolates oxidized disaccharides (sucrose, lactose, maltose) and sugar alcohols (manitol, sorbitol and dulcitol) within 3–5 days (Table 3). The oxidation reaction was indicated by a change in colour. The results revealed a change from a blue to a yellow colour, indicating the oxidization of sugars by bacterial isolates. Therefore, all groups of R. solanacearum isolates belong to biovar III, as shown in Table 3. On the other hand, all the control plates of different sugars and sugar alcohols remain unchanged.
Differentiation of Ralstonia solanacearum isolates groups into biovars and races used in this study.
| Isolate Group |
Locations | Host | No. of isolates | Utilization of carbohydrates | Biovars | Races | |||||
| Dextrose | Maltose | Lactose | Sorbitol | Manitol | Dulsitol | ||||||
| RsB-1 | Panchagarh | Eggplant | 4 | + | + | + | + | + | + | III | I |
| RsB-2 | Rangpur | Eggplant | 4 | + | + | + | + | + | + | III | I |
| RsB-3 | Jessore | Eggplant | 4 | + | + | + | + | + | + | III | I |
| RsP-1 | Nilphamari | Potato | 4 | + | + | + | + | + | + | III | III |
| RsP-2 | Munshigonj | Potato | 4 | + | + | + | + | + | + | III | III |
| RsP-3 | Jamalpur | Potato | 4 | + | + | + | + | + | + | III | III |
| RsP-4 | Panchagarh | Potato | 4 | + | + | + | + | + | + | III | III |
There is no biochemical test for race identification of R. solanacearum. The races of R. solanacearum were identified by pathogenicity tests in a wide host range. The result of the pathogenicity test showed that all four groups of R. solanacearum isolates obtained from eggplant tested in the study were able to cause wilt symptoms in inoculated eggplant, tomato and chilli plants (Table 3). Therefore, all groups of R. solanacearum isolates causing the bacterial wilt of eggplant belong to race 1. On the other hand, none of the groups of R. solanacearum isolates obtained from potato tested in the study was able to cause wilt symptoms in inoculated eggplant, tomato and chilli plants indicating a narrow host range, but the isolates produced the wilt symptom in potato seedlings. Therefore, all groups of R. solanacearum isolates causing the bacterial wilt of potato collected from three selected growing areas belong to race 3.
3.4 RAPD analyses of R. solanacearum isolates
To assess the genetic diversity analyses of R. solanacearum isolates collected from eggplant and potato from different growing areas, we have chosen a total of 28 isolates out of 95 that representing 4 isolates from each area, which were grouped into 7 samples based on the place where the isolates had been collected. Although the number of isolates is low for analyses of genetic diversity, it will be a first step to find out the suitability of RAPD markers for genetic diversity analyses of R. solanacearum in Bangladesh. Among the 15 primers initially tested, 67AB10G07, OPA02, OPB08 and OPB17 produced a higher number of amplification products with high intensity, minimal smearing, and good resolution (Fig. 2A–D). Four primers generated various banding patterns, the number of bands ranged from 14 to 21. The primer 67AB10G07 amplified a maximum number of polymorphic bands. The size of the PCR amplification products ranged from 100 to 1400 bp. The four primers generated 17.5 scorable bands per primer and 17 polymorphic RAPD markers per primer (Table 4). The RAPD primers generated 70 bands, of which 68 bands were polymorphic (97.06%) and two (2.94%) were monomorphic amongst the seven group of Ralstonia isolates (Table 4). Strong and weak bands were produced in the RAPD reactions.
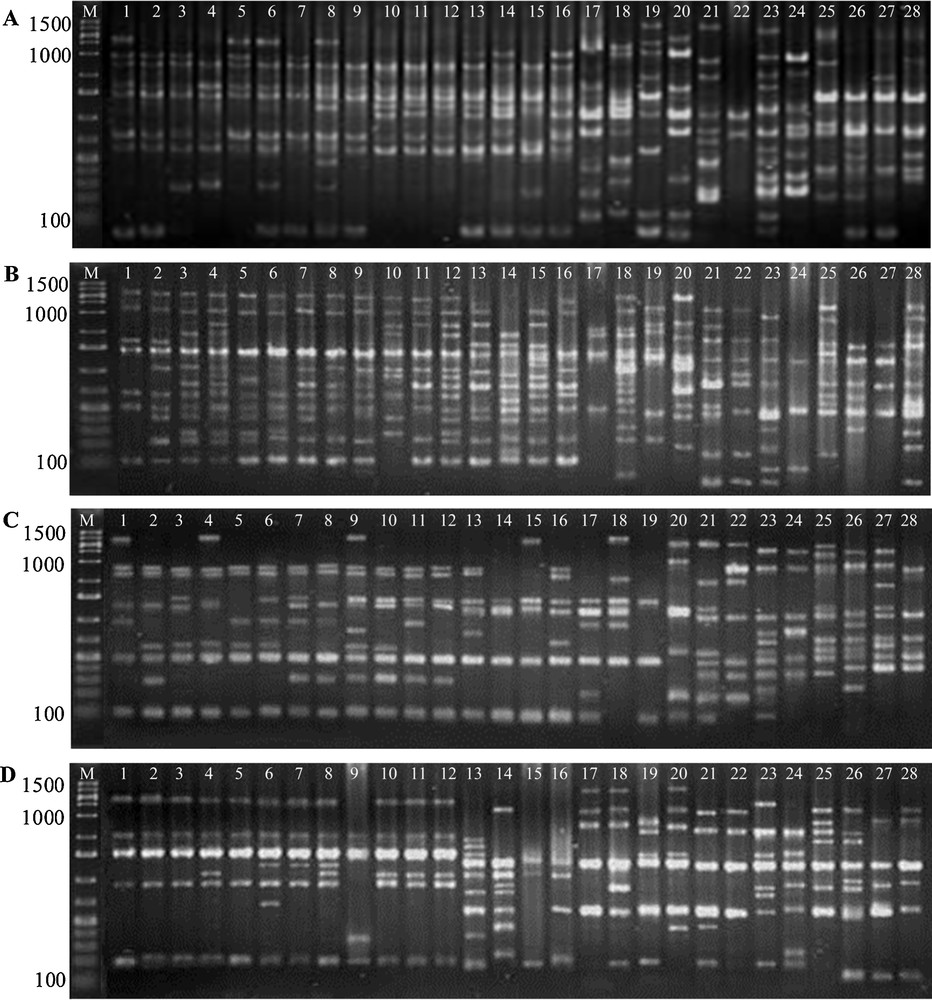
RAPD amplification profiles of seven isolate groups (total of 28 isolates) of R. solanacearum using primers A. 67AB10G07, B. OPA02, C. OPB08 and D. OPB17. M: 20 bp ladder, Lane 1-4: RsB-1, Lane 5-8: RsB-2, Lane 9-12: RsB-3, Lane 13-16: RsP-1, Lane 17-20: RsP-2, Lane 21-24: RsP-3, Lane 25-28: RsP-4 isolates groups of R. solanacearum.
RAPD primers with corresponding bands scored and their size range together with polymorphic bands observed in seven isolate groups of R. solanacearum.
| Primer | Sequences (5′-3′) | No. of bands scored | Size ranged (bp) | No. of polymorphic bands | Polymorphic loci (%) |
| 67AB10G07 | TTGGCACGGG | 21 | 100-1400 | 21 | 100.00 |
| OPA02 | TGCCGAGCTG | 17 | 100-1400 | 15 | 88.24 |
| OPB08 | GTCCACACGG | 14 | 100-1300 | 14 | 100.00 |
| OPB17 | GACCGCTTGT | 18 | 100-1400 | 18 | 100.00 |
| Total | 70 | 68 | 388.24 | ||
| Average | 17.5 | 17 | 97.06 |
3.5 Intra-isolate similarity indices
The highest intra-isolate similarity indices (Si) value was found in RsB-1 isolate (86.35%), followed by that found in RsB-3 and RsB-2. The intra-isolate similarity indices (Si) value was 56.59% in RsP-2 (Table 5). The isolates showing higher intra-isolate similarity and lower frequency of polymorphic loci had less heterozygosity as compared to those showing lower intra-isolate similarity. In other words, isolates with lower intra-isolate similarity formed a less homogenous group.
Band sharing based on percentage similarity indices between individuals of seven isolate groups of R. solanacearum.
| Isolates | Band sharing values (%) | ||||
| 67AB10G07 | OPA02 | OPB08 | OPB17 | Average | |
| RsB-1 | 81.90 | 82.90 | 85.16 | 95.45 | 86.35 |
| RsB-2 | 70.79 | 91.33 | 83.21 | 79.37 | 81.18 |
| RsB-3 | 84.61 | 79.01 | 91.14 | 79.39 | 83.54 |
| RsP-1 | 82.76 | 74.99 | 75.78 | 64.80 | 74.58 |
| RsP-2 | 64.49 | 53.12 | 40.97 | 67.77 | 56.59 |
| RsP-3 | 38.68 | 63.67 | 61.90 | 68.95 | 58.30 |
| RsP-4 | 41.56 | 72.01 | 79.46 | 77.34 | 67.59 |
3.6 Frequency of polymorphic loci
The number and proportion of polymorphic loci was the highest in RsP-3 (they were 41 and 58.57%, respectively, Table 6). The highest level of Nei's [35] gene diversity value (0.2132) was also found in these isolates. On the other hand, a minimal proportion of polymorphic loci (22.86%) and Nei's (1973) gene diversity value was observed in RsB-3 (0.1350). With the lowest intra-variety similarity (Si) value (56.59%), the highest gene diversity value (h = 0.2132), the highest Shannon Information index (I = 0.3186), and the highest proportion of polymorphic loci (58.57%), RsP-3 was diversified among the seven isolates. On the other hand, RsB-3 was the least diversified, as it comprises the highest value of intra-variety similarity (Si), the lowest gene diversity value, the lowest Shannon Information index (I), and the lowest proportion of polymorphic loci (22.86%), as shown in Table 4.
Genetic variation of the seven groups of R. solanacearum isolates as measured by the number and the proportion of polymorphic bands along with gene diversity and Shannon's index.
| Isolates | No. of polymorphic loci | Proportion of polymorphic loci (%) | Gene diversity | Shannon's Information index (I) |
| RsB-1 | 17 | 24.29 | 0.0911 | 0.1351 |
| RsB-2 | 17 | 24.29 | 0.1003 | 0.1459 |
| RsB-3 | 16 | 22.86 | 0.0928 | 0.1350 |
| RsP-1 | 28 | 40.00 | 0.1455 | 0.2171 |
| RsP-2 | 39 | 55.71 | 0.1963 | 0.2958 |
| RsP-3 | 41 | 58.57 | 0.2132 | 0.3186 |
| RsP-4 | 26 | 37.14 | 0.1390 | 0.2063 |
3.7 Genetic distance
The values of pairwise comparisons of Nei's [34] genetic distance among the seven group of R. solanacearum isolates ranged from 0.0357 to 0.4293. Comparatively, a higher genetic distance (0.4293) was observed between RsB-2 and RsP-4 isolates than with other combinations. The lowest genetic distance (0.0357) was found in RsB-1 vs RsB-2 (Table 7).
Nei's (1972) genetic identity (above diagonal) and genetic distance (below diagonal) values between the seven groups of isolates of Ralstonia solanacearum.
| Isolates | RsB-1 | RsB-2 | RsB-3 | RsP-1 | RsP-2 | RsP-3 | RsP-4 |
| RsB-1 | **** | 0.9650 | 0.9369 | 0.8142 | 0.6847 | 0.6797 | 0.6658 |
| RsB-2 | 0.0357 | **** | 0.9311 | 0.8421 | 0.7118 | 0.6987 | 0.6510 |
| RsB-3 | 0.0652 | 0.0714 | **** | 0.8294 | 0.7072 | 0.6968 | 0.6618 |
| RsP-1 | 0.2055 | 0.1719 | 0.1871 | **** | 0.7865 | 0.7214 | 0.7128 |
| RsP-2 | 0.3788 | 0.3400 | 0.3465 | 0.2401 | **** | 0.8445 | 0.7546 |
| RsP-3 | 0.3861 | 0.3585 | 0.3613 | 0.3266 | 0.1690 | **** | 0.8562 |
| RsP-4 | 0.4068 | 0.4293 | 0.4129 | 0.3386 | 0.2815 | 0.1552 | **** |
3.8 Dendrogram showing relationship among the R. solanacearum isolates
The dendrogram constructed based on Nei's [34] genetic distance using the Unweighted Pair Group Method of Arithmetic Means (UPGMA) indicated a segregation of seven groups of R. solanacearum isolates into two main clusters: cluster 1 and cluster 2 (Fig. 3). The isolates RsB-1, RsB-2, RsB-3 and RsP-1 grouped in cluster І, while RsP-2, RsP-3 and RsP-4 gathered in cluster ІІ. Cluster І was divided into two groups, RsP-1 alone formed one group and the isolates collected from brinjal (RsB-1, RsB-2 and RsB-3) formed another group. Again, cluster ІІ formed two groups, in which RsP-2 alone belonged to one group. RsP-3 and RsP-4 belonged to another group. This study indicates that RsB-1 and RsB-2 showed a closer relationship, with a minimal genetic distance (0.0357) in cluster І. The highest genetic distance (0.4293) was found between RsB-2 and RsP-4.

Dendrograms constructed by the unweighted pair group method with arithmetic mean (UPGMA) showing dissimilarity among random amplified polymorphic DNA (RAPD) haplotypes of R. solanacearum isolate groups from eggplant (RsB-1 to RsB-3) and from potato (RsP-1 to RsP-4). Clusters showing the genetic distance among seven isolate groups of R. solanacearum with the relationship among biovars, races, and their geographical distribution.
4 Discussion
In this study, a significant variation was observed in bacterial wilt incidence and severity in different growing areas of eggplant and potato, which primarily indicate a variation in the R. solanacearum population, probably due to the differences in host responses as well as to the genetic diversity in pathogen populations. Differences of wilt incidence and severity were reported due to the diversity of host plants affected by this pathogen, phenotype and genotype of R. solanacearum, its wide geographical distribution, and the range of environmental conditions conducive to bacterial wilt [36]. However, the differentiation of biovars of R. solanacearum isolates obtained from eggplant and potato based on the utilization of carbohydrates revealed that R. solanacearum isolates infecting eggplant and potato belong to same biovar III. Similar observations were reported previously by Hayward [37], He et al. [8] and Kumar et al. [38]. They observed that biovar III oxidizes both disaccharides and hexose alcohols, whereas biovar I oxidize hexose alcohols but not disaccharides, biovar II oxidizes only disaccharides and biovar IV oxidizes only alcohols. RAPD data revealed that all groups of R. solancearum isolates belonging to biovar III are distributed into two main clusters. Previously, it was also observed biovar 3 was more heterozygous as compared to biovar 4 [39]. Horita and Tsuchiya [40] reported that biovar III strains have low average similarity and fall into five groups. Moreover, Poussier et al. [41] have demonstrated that biovar III strains cluster into two groups, which revealed their higher level of genetic dissimilarity.
Identification of races by pathogenicity test on tomato, potato, eggplant and chilli showed that R. solanacearum isolates obtained from eggplant belong to race 1, while isolates of R. solanacearum collected from potato belong to race 3. Denny and Hayward [42] identified the races of R. solanacearum by host range. The findings of the present study are also supported by Buddenhagen and Kelman [43], who divided R. solanacearum into three races. Race 1 infects many solanaceous plants such as brinjal, tomato, tobacco, pepper and other plants, including some weeds. However, race 2 causes wilt of triploid banana (Musa spp.) and Heliconia spp. Race 3 affects potato and tomato but is weakly virulent on other solanaceous crops. Later, Aragaki and Quinon [44] reported race 4 from infected ginger in the Philippines. He et al. [8] reported race 5 from mulberry in China. Five races have been described so far, but they differ in host range, geographical distribution, and ability to survive under different environmental conditions [45]. Patrice (2008) reported that R. solanacearum was initially subdivided into races and biovars based on variability in host range. He added that five races have been identified within the species. Strains of R. solanacearum have also been divided into five host-specific races by Pradhanang et al. [46]. The dendrogram based on Nei's [34] genetic distance using the Unweighted Pair Group Method of Arithmetic Means (UPGMA) indicated the segregation of 28 isolates of R. solanacearum grouped into seven, which were clustered into two major clusters: cluster 1 and cluster 2. The isolates RsB-1, RsB-2, RsB-3 and RsP-1 grouped in cluster І while RsP-2, RsP-3 and RsP-4 in cluster ІІ. Although, it is usual that the isolates from the same host are distributed in the same cluster, one group of isolates, RsP-1, obtained from potato alone, formed a cluster with the isolates collected from brinjal (RsB-1, RsB-2 and RsB-3). These results indicate that this group of isolates may be the clone of the field isolates introduced in that area initially. Again, cluster ІІ is comprised of two groups, where RsP-2 alone belongs to one group and RsP-3 and RsP-4 belongs to another group. The absence of association between the genotypic and pathotypic groupings was also proposed by Leung et al. [47]. They assumed that:
- • the pathogen population is strongly selected by host genotype;
- • the mutation rate of the pathogen is variable;
- • or the pathotype, as defined by interactions with specific host cultivars, is not the main unit of pathogen evolution.
This study indicates that RsB-1 and RsB-2 showed closer relationship with minimal genetic distance (0.0357) in cluster 1 and RsB-2 and RsP-4 were distantly related in cluster 2 with a maximum genetic distance (0.4293).
In the present study, four primers out of 15 generated various banding patterns with 97.06% polymorphic and 2.94% monomorphic bands amongst the seven groups of R. solanacearum isolates. All these primers produced a highly polymorphic banding pattern, confirming the extreme heterogeneity within this field population of this soil-borne bacterium. Although it is not known when exactly this bacterium was introduced in the country, it was surprising to find so much diversity. Genetic diversity was so high that maximum similarity only to the extent of 41% could be obtained among the isolates. In fact, the fragment size frequency of 58.57% suggested that they represent regions that are highly variable from one isolate to another. A least variability in terms of band frequency suggests that those particular regions of the R. solanacearum genome possibly play a critical role in its survival and are, therefore, resistant to change. The highest level of Nei's [35] gene diversity value (0.2132) was also found in these isolates. On the other hand, the minimal proportion of polymorphic loci (22.86%) and Nei's (1973) gene diversity value was observed in RsB-3 (0.1350). With the lowest intra-variety similarity (Si) value (56.59%), the highest gene diversity value (h = 0.2132), the highest Shannon's Information index (I = 0.3186), and the highest proportion of polymorphic loci (58.57%), RsP-3 was diversified among the seven groups of isolates. On the other hand, RsB-3 was the least diversified, as it comprises the highest value of intra-variety similarity (Si), the lowest gene diversity value, the lowest Shannon's Information index (I) and the lowest proportion of polymorphic loci (22.86%). R. solanacearum is a heterogeneous complex species of soil-borne bacterium which is extreme in terms of variability in races, biovars [37,48], with a wide host range. The non-availability of durable resistance along with its complex races and biovars together indicate the existence of high genetic diversity within this species. Recently, the entire genome of R. solanacearum (strain GMI 1000) has been sequenced [49], and the existence of several mobile elements, prophages, insertion sequences, transposons and the like in the genome provides clues for genetic instability in this bacterium, observed earlier by several workers [39,40,50–52]. In addition, there are several loci in the genome that are considered recombinational hot spots, thus suggesting rapid genome evolution. A hypothesis is also consistent with the ability of the bacterium to be naturally transformed by the acquisition of genetic material from the environment and insertion of the incoming DNA into its genome [53,54], indicating that the genome sequence published by the group might represent a single snapshot of a structure that is variable from isolate to isolate and within derivatives of the same isolate. In fact, R. solanacearum is the only bacterial plant pathogen where horizontal gene transfer even from archaebacteria has been reported [55]. The combination of lateral gene transfer, sequence amplification, gene inactivation and ultimately mutation may be the reasons for the observed genetic diversity in the natural population of this bacterium. Our data indicated that R. solanacearum is capable of creating very high levels of genetic diversity among the isolates obtained from different regions, irrespective of the hosts that were clustered at a low level of similarity index. This explains the fact that the large variability observed in this population of R. solanacearum can also be related to its soilborne nature. The pathogen has to cope with the variable environment in the soil during its saprophytic survival [11]. Thus, soil properties may play an important role in genetic differentiation, as has been demonstrated for other soilborne bacterial species [3]. Ecologists and population geneticists have long suspected that the structure of the environment is connected with the maintenance of diversity in micro-organisms [56]. This bacterium is well known to survive under diverse ecological conditions, interacting with a milieu of biotic and abiotic factors. That may be the reason for the evolution and maintenance of higher variability in this bacterium.
To our knowledge, this is the first report of genetic diversity assessment among R. solanacearum field isolates collected from different hosts and locations in Bangladesh. However, for getting a better correlation between isolates and hosts source, more isolates are needed from various host plants. In spite of these facts, at least it may be concluded from the present study that RAPD markers may be among the more sensitive, simple, efficient tools for genetic diversity analysis among and within R. solanacearum isolates and effectively trace their genetic relationships. However, using larger numbers of bacterial isolates and more primers for RAPD along with the use of the other molecular markers in analysing the genetic diversity of R. solanacearum will unravel the complex population structures of this soil-borne bacterium in the country. The best management strategy of bacterial wilt is host resistance. Information and knowledge on the existence of variability in the local pathogen population and host range determination will be the first step when setting up a breeding for resistance strategy. Finally, the information provided by this study about the diversity of R. solanacearum strains will be useful for starting and optimization of pathogen population analyses and breeding programs in Bangladesh.
Acknowledgement
This research was partly supported by a USDA grant to Prof. Dr. M. Bahadur Meah and a UGC-BAURES research grant to Prof. Dr. Md. Rashidul Islam.


