1 Introduction
Rice is an important cereal crop in the world and provides more than 21% of the food for the world population and up to 76% of the caloric intake in Southeast Asia [1,2]. Basmati is one of the premium rice groups, cultivated in the northwestern plains of India and eastern Pakistan. There is a marked increase in demand for Basmati rice worldwide for its unique aroma, cooking, and eating qualities [3]. In India, more than 60% of the total Basmati rice is grown in Haryana, and thus the latter is the major Basmati rice cultivating state. India is the leading exporter of Basmati rice and exports US$ 4.86 billion of Basmati rice annually [4]. Aroma, non-stickiness of cooked rice and elongation upon cooking are some of the important traits of Basmati rice. Bradbury et al. [5] showed that a functional BADH2 enzyme inhibits the biosynthesis of 2-acetyl-1-pyrroline, which is a major component of aroma. A fully functional copy of the gene encoding BAD2 is present in non-fragrant varieties, whereas fragrant varieties possess a copy of the gene containing an eight-base pair deletion resulting in a frame shift mutation disabling the BADH2 enzyme. Studies by Kovach et al. [6] also confirmed that BADH2 is the major genetic determinant of fragrance in rice. Amylose content and alkali spread value (ASV) are the most important constitutional indices related to cooking and processing quality of rice [7,8].
BB disease is one of the most devastating diseases effecting Basmati rice acreage in India. BB is caused by the Gram-negative proteobacterium, Xanthomonas oryzae pv. Oryzae, and causes yield losses ranging from 74% to 81% in severe conditions, depending on the stage of the crop, the cultivar's susceptibility, and the environmental conditions [9,10]. Effective control of disease is limited by the use of chemical control measures and the health issues posed by them [11,12]. The development of varieties carrying resistance genes has been considered to be the most effective way to control the BB disease [13–16]. To date, at least 38 BB resistance genes [27 dominant (Xa1, Xa2, Xa3, Xa4, Xa7, Xa10, Xa11, X12, Xa14, Xa16, Xa17, Xa18, Xa21, Xa22 (t), Xa23, Xa25 (t), Xa26, Xa27, Xa29, Xa30, Xa30 (t), Xa31 (t), Xa32 (t), Xa34, Xa35 (t), Xa36 (t), Xa38), and 11 recessive (xa5, xa5(t), xa8, xa13, xa15, xa19, xa20, xa24, xa28 (t), xa31 and xa32)] conferring host resistance against various strains of Xoo have been identified [17,18].
Molecular breeding involving MAS addresses the limitations of conventional breeding and allows pyramiding of multiple resistance genes into a single cultivar, thus providing an effective solution to resistance breakdown. Using the gene pyramiding approach, improved rice cultivars with broad spectrum and durable BB resistance have been developed by combining multiple resistance genes [13–16,19–22]. A marker-assisted background selection approach can recover up to 99% of the recurrent-parent genome in just three backcross cycles, whereas conventional breeding takes up to six backcrosses to recover 99% of the recurrent parent genome [23]. BB resistance genes xa13 and Xa21 have been introgressed into ‘Pusa Basmati 1′ through MAS coupled with phenotypic selection for agronomic, grain, and cooking quality traits [22]. One of the improved genotypes was released as ‘Improved Pusa Basmati 1′ for commercial cultivation in 2007, and is one of the first products of MAS to be released in India [24]. In the present study, we employed MAS along with stringent phenotypic selection for the introgression of three BB resistance genes, viz., Xa21, xa13, and xa5 into Basmati variety CSR-30. This work will facilitate future efforts to transfer combinations of BB resistance genes into other preferred Basmati rice cultivars from non-Basmati rice cultivars without compromising the Basmati quality characters.
2 Material and methods
2.1 Plant material
The plant material consisted of the Basmati rice (susceptible) variety CSR-30 as a recurrent parent, the (resistant) IRBB-60 variety as a donor parent having BB resistance genes xa5, xa4, xa13, and Xa21. Crosses were made between CSR-30 and IRBB-60, and F1 plants thus obtained were backcrossed with CSR-30. Among the BC1F1 population, the foreground selection was performed using polymerase chain reaction (PCR)-based sequence tagged sites (STS) markers linked to the three BB resistance genes xa5, xa13, and Xa21. The BC1F1 genotypes were advanced to BC1F2 and subjected to foreground selection followed by BB incidence analysis on artificial inoculation and background analysis. A similar strategy was used in the BC2F1 to obtain BC3F1 populations from which three-gene positive genotypes were selected based on molecular marker analysis and assessed for agronomic performance and disease response at the tillering stage. The BC3F1 plants having the three BB resistance genes were selfed and BC3F2 seeds thus obtained were used for grain quality evaluation.
2.2 DNA marker analysis and PCR amplification
Mini-scale genomic DNA isolation was carried out using the CTAB extraction method of Murray and Thompson [25] as modified by Saghai-Maroof et al. [26] and Xu et al. [27]. Three STS markers, viz., pTA248, RG136 and RG556 linked to the BB resistance genes, Xa21, xa13 and xa5, respectively, were used to confirm the presence of these resistance genes at each backcross generation. The pTA248 marker is 0.2 cM from Xa21 [28], the RG136 marker is 3.8 cM from xa13 [29], and the RG556 marker is 1.7 cM from xa5 [30] (Table 1). PCR was carried out in 20-μl reaction mixtures containing 50 ng of genomic DNA, 2 units of Taq DNA polymerase, 1X PCR Buffer (10 mM Tris HCL, 1.5 mM MgCl2), 100 μM each of dNTPs, and 10 μM of each primer. The template DNA was initially denatured at 94 °C for 5 min, followed by 30 cycles of PCR amplification with the following conditions: 30 s of denaturation at 94 °C, 1 min of annealing at 55 °C, and 1 min of primer extension at 72 °C followed by final extension at 72 °C for 10 min. The amplified product of pTA248 was electrophoretically resolved on 1.5% agarose gel containing 0.5 μg/ml of ethidium bromide in a 1.0 X TBE buffer and visualized under UV light. For the amplified products of RG136 and RG556, 5 μl of the PCR product were used for gel electrophoresis to check DNA amplification. The remaining PCR product was used for restriction digestion. The reaction mixture used for the digestion of the PCR product with the respective restriction enzyme consisted of 0.3 μl (10 U/μl) of restriction enzyme Hinf1 for RG136 amplicons and Dra1 for RG556 amplicons, 2.5 μl of 10 X restriction buffer, 7.2 μl of sterile distilled water and 15 μl PCR product to make a final volume of 25 μl [31]. The reaction mixture was incubated for 5 h at 37 °C, and the products of restriction digestion were separated by gel electrophoresis (2.0% agarose) and visualized under UV light. MAS was also carried out for fgr gene for aroma located on the short arm of rice chromosome 8 using primer BAD2 [5] (Table 1).
Molecular markers used for marker-assisted selection of BB resistance genes and aroma gene.
| Molecular marker | R genes for BB/fgr for aroma | Chromosome | primer sequences (5′–3′) | Reference |
| pTA248 (0.2cM) |
Xa21 | 5 | F:5′AGACGCGGAAGGGTGGTTCCCGGA3′ R:5′AGACCGGTAATCGAAAGATGAAAA3′ |
Yoshimura et al. [30] |
| RG136 (3.8cM) |
xa13 | 8 | F:5′TCCCAGAAAGCTACTACAGC3′ R:5′GCAGACTCCAGTTTGACTTC3′ |
Zhang et al. [29] |
| RG556 (1.7cM) |
xa5 | 11 | F:5′TAGCTGCTGCCGTGCTGTGC3′ R:5′AATATTTCAGTGTGCATCTC3′ |
Ronald et al. [28] |
| BAD2 | fgr | 8 | F:5′TTGTTTGGAGCTTGCTGATG3′ F:5′CTGGTAAAAAGATTATGGCTTCA3′ R:5′CATAGGAGCAGCTGAAATAATACC3′ R:5′AGTGCTTTACAAAGTCCCGC3′ |
Bradbury et al. [5] |
For background analysis, 131 polymorphic SSR markers out of the 428 tested were used for the assessment of the relative contribution of the two parental genomes to the pyramided genotypes. The computer package NTSYS-PC [32] was used for cluster analysis. Graphical genotypes (GGT) Version 2.0 [33] software programme was used for the assessment of the genomic contribution of the parent in the selected genotypes based on SSR marker data.
2.3 BB inoculation and resistance evaluation
Plants selected based on molecular marker analysis from the BC1F2 and BC3F1 generation carrying resistance genes (Xa21, xa13 and xa5) individually and in combinations, along with the control, were inoculated with the predominant Xoo isolate prevalent in Kaul, Haryana, using a bacterial suspension of 109 cells/ml [34]. The plants were clip inoculated at the maximum tillering stage. Inoculation of the leaf blades was done by clipping with scissors at 3 cm below the leaf tips. On average, five leaves per plant were inoculated, and the disease incidence using the 0–5 scale was measured 14 days after inoculation.
2.4 Evaluation of quality traits
Plant height, effective tillers per plant, panicle length, filled grains per panicle, 1,000 grain weight and grain yield were recorded in the rice genotypes pyramided with three resistance genes in BC1F2 and BC3F1 generation. The pyramided BC1F2 and BC3F1 genotypes were grown during the kharif season of 2012 and 2014 in a net house at CCSHAU, Hisar, and in field at the Regional Rice Research Station, Kaul. Harvested seeds from BC3F1 three-gene-pyramided genotypes were further analysed for physicochemical characteristics like kernel dimensions of uncooked and cooked rice, aroma, ASV and amylose content. For testing aroma, one gram milled rice kernels were soaked in 10 ml of 1.7% KOH at room temperature in covered Petri plates for 10 min [45]. The amylose content of the pyramided genotypes was estimated according to Perez and Juliano [35]. ASV was assayed according to the method of Little et al. [36].
3 Results
3.1 Foreground and background selection for BB resistance genes
In BC1F2 population, plants with single and with three resistance genes were identified. The presence of BB resistance genes, viz., Xa21, xa13, and xa5 in the BC1F2 genotypes were determined by respective linked marker and 10/230 genotypes had all the three BB resistance genes. Among resistant genotypes with different gene combinations, 44 were homozygous for Xa21 (i.e. Xa21Xa21), six genotypes were homozygous for xa13 (i.e. xa13xa13), nine genotypes were homozygous for xa5 (i.e. xa5xa5) and the remaining plants were heterozygous, either for Xa21 or xa13 or xa5. A total of 104 polymorphic SSR markers spanning uniformly across all the rice chromosomes were used for background selection among the ten three-gene-pyramided genotypes. RPG recovery ranged from 44.1% to 78.9%. Three out of ten plants that showed RPG recovery above 70% were backcrossed with CSR-30, and BC2F1 generation was obtained, which was further advanced to BC3F1. Of the 112 BC3F1 plants, 15 genotypes were positive for all the three BB resistance genes using the linked markers (Fig. 1). In BC3F1 generation, background selection was performed using a total of 131 polymorphic SSR markers. The similarity indices between different genotypes ranged from 0.02 to 0.97 across all the genotypes. The donor parent (IRBB-60) and recurrent parent (CSR-30) had low genetic similarity coefficient and bifurcated at a coefficient value of 0.09. Basmati and non-Basmati genotypes are highly differentiated genetically, as evident from the cluster pattern. The maximum genetic similarity coefficient for the three-gene-pyramided BC3F1 rice genotypes with respect to recurrent parent CSR-30 was observed in IC-R28 (0.97) followed by IC-R68 (0.94), IC-R32 (0.93) and IC-R42 (0.93). UPGMA cluster tree was divided into two groups (Fig. 2). The recipient parent CSR-30 and all the BC3F1rice plants positive for Xa21, xa13 and xa5 BB resistance genes fell in one group with two major sub-groups. Sub-group I consisted of recipient parent CSR-30 and two genotypes, IC-R28 and IC-R32 with maximum similarity to CSR-30. Sub-group II consisted of IC-R42 and IC-R68 with close similarity to CSR-30 and the remaining eleven genotypes. In BC3F1, RPG recovery ranged from 72.9% to 97.1%. On BB resistance genes carrier chromosomes, 6 polymorphic markers were found on chromosome 5, 8 on chromosome 8 and 5 on chromosome 11, respectively. Maximum donor parent genomic regions among the three carrier chromosomes were present on chromosome 8 and chromosome 11 flanking the genes xa13 and Xa21, as a result of linkage drag (Fig. 3). Background analysis conferred that IC-R28, IC-R68, IC-R32 and IC-R42 had the least introgression of donor segments. These four three-gene-pyramided genotypes with RPG above 89% were selfed and BC3F2 seeds were harvested and analysed for grain and cooking quality traits. Screening for BB resistance, and agronomic performance resulted in a cost-effective selection process due to a reduction in the number of plants subjected to foreground selection. The molecular analysis with BAD2 primer combination specific for aroma (fgr) produced a 257 bp fragrance specific allele in all 15 recombinants and CSR-30, whereas IRBB-60 produced a 355 bp non-fragrance allele (Fig. 1d).
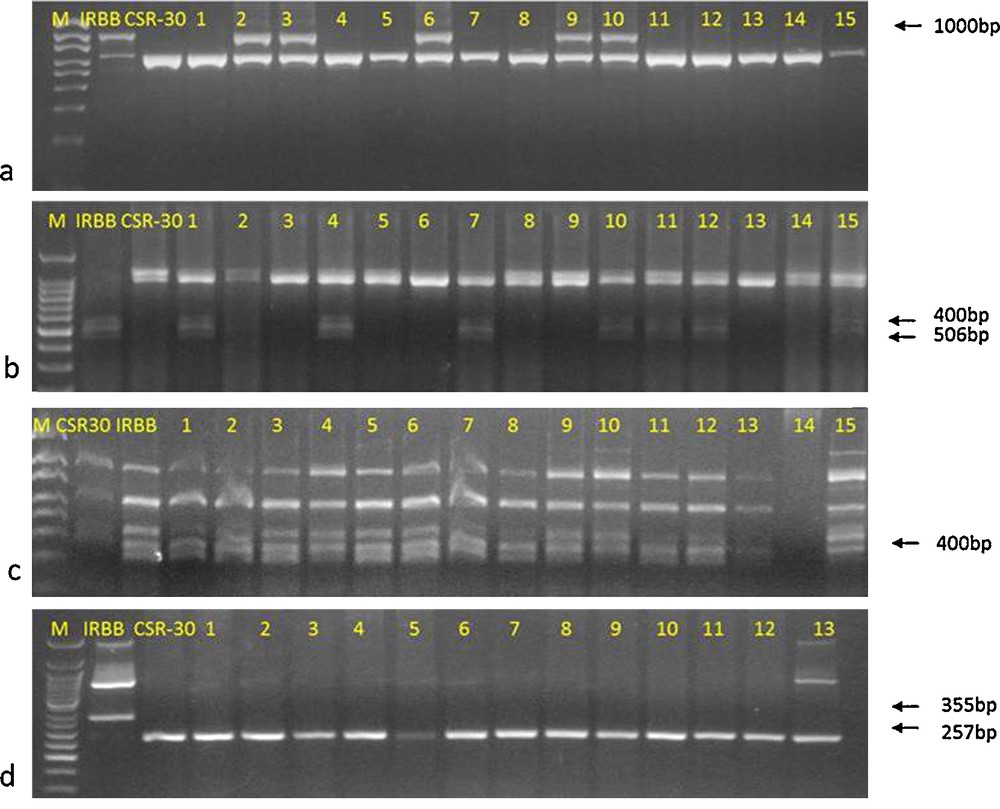
Marker-assisted selection in BC3F1 genotypes from cross CSR-30 × IRBB-60 for (a) Xa21 using pTA248 (b) xa13 using RG136 (c) xa5 using RG556, and (d) fgr using BAD2 markers.
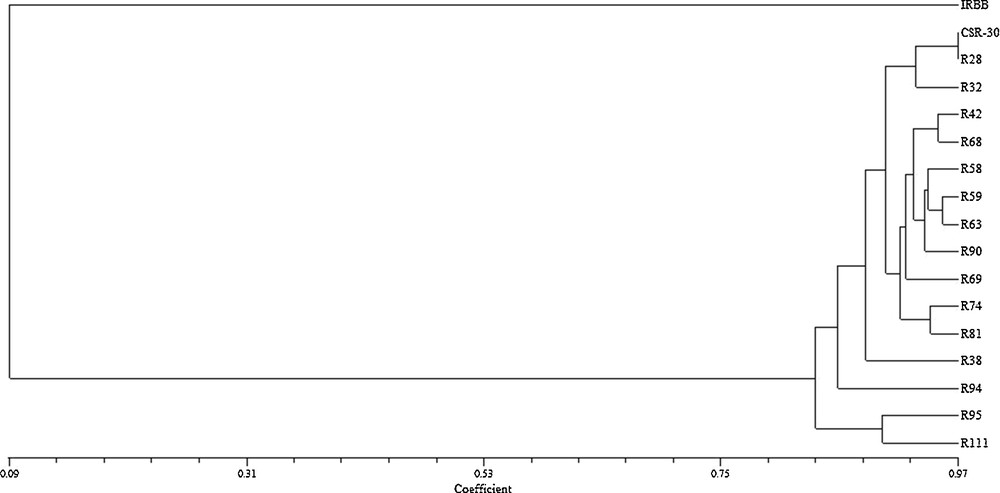
Dendrogram (NTSYS-pc, UPGMA) showing the genetic similarity among the BC3F1 three-gene-pyramided genotypes and the parental rice genotypes based on SSR diversity data at 428 loci.
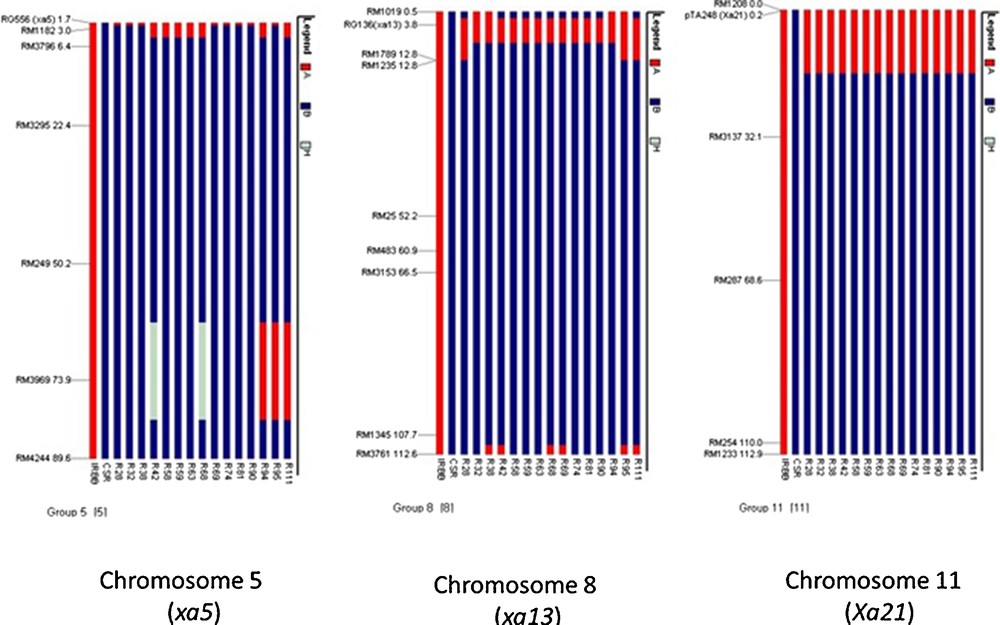
Demonstration of the donor parent (IRBB-60) and recurrent parent (CSR-30) genome on chromosomes 5, 8 and 11 (having BLB resistance genes xa5, xa13 and Xa21) in the 15 three-gene-pyramided BC3F1 genotypes.
3.2 Evaluation of BB resistance
The BC3F1 pyramided genotypes were evaluated for their resistance to BB under glass house conditions using the Xanthomonas oryzae strain isolated from the BB infected fields of RRS, Kaul, CCSHAU, Hisar. The three-gene-pyramided BC3F1 genotypes with mean lesion length of 0.4 cm, were found to be equally effective against the virulent Xoo strain as the donor parent IRBB-60 with a mean lesion length of 0.5 cm. Also, the genotypes having either Xa21 (mean lesion length of 1.2 cm) or xa5 (mean lesion length of 1.1 cm) resistance genes alone were found to be resistant or moderately resistant to the BB disease. However, genotypes with xa13 gene (mean lesion length of 4.8 cm) alone were found to be susceptible to BB disease. The pyramided genotypes with three genes had a higher level of resistance and a broader resistance spectrum than parental genotypes or genotypes with a single gene. The results indicated that the genes in combinations were more effective against the pathogen than a single gene (Fig. 4).
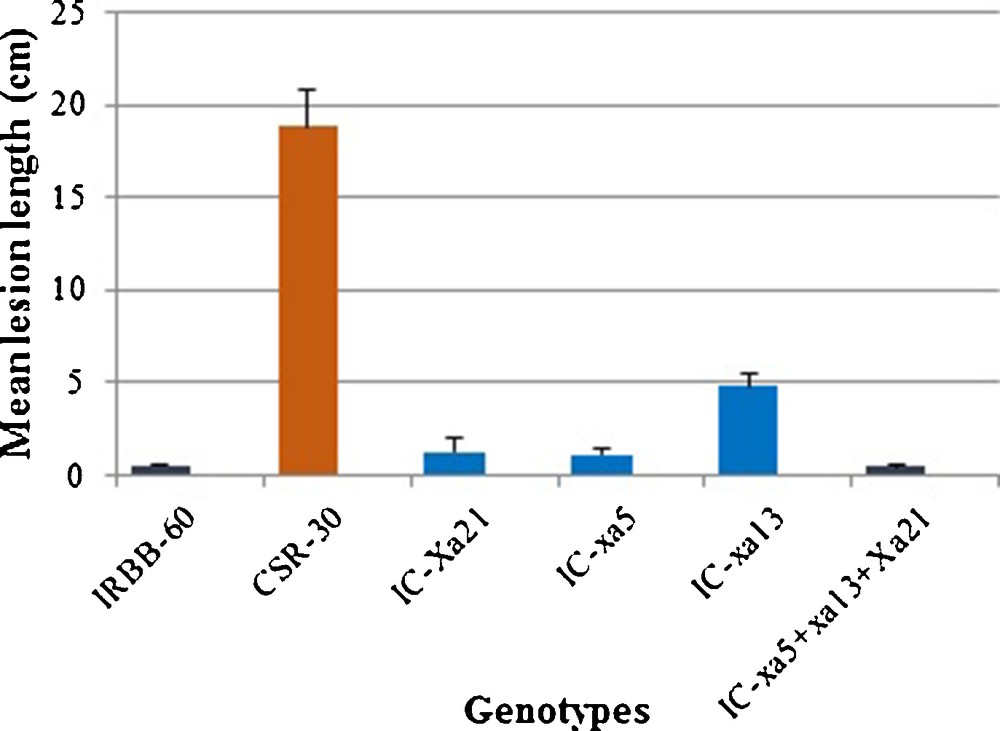
Disease reaction of the donor parent (IRBB-60), the recurrent parent (CSR-30), and the pyramided BC3F1 genotypes (carrying one to three BB resistance genes).
3.3 Agronomic performance and quality traits
Significant variation was observed among the pyramided lines and parental rice genotypes for plant height, tillers per plant, panicle length, filled grains/panicle, and grain yield per plant (Table 2). Most of the three gene-pyramided BC3F1 genotypes were similar or superior to the recurrent parent CSR-30 for the agronomic traits. In the pyramided BC3F1 genotypes, the plant height ranged from 130.5 to 154 cm, whereas the plant height of the recurrent parent CSR-30 was 164.5 cm. The three gene-pyramided BC3F1 genotypes had plant height intermediate between the those of the two parents. The fifteen pyramided BC3F1 genotypes had 1000 grain weight greater than the recurrent parent CSR-30 (21.2 g). The genotypes with maximum recovery of RPG, IC-R28 (31.11 g/plant), IC-R32 (33.22 g/plant), IC-R42 (36.78 g/plant) and IC-R68 (33.38 g/plant) showed grain yield/plant higher than that of the recurrent parent CSR-30 (31.08 g/plant).
Agronomic traits, BB reaction and % recurrent parent genome (RPG) recovery in the three-gene-pyramided BC3F1 genotypes.
| S. No. | Plant No. | PH (cm) | NT | 1000 GW (g) | PL(cm) | FG/P | Awns | Y/P (g) | BB lesion length (cm) | RPG (%) |
| 1 | IRBB-60 | 109 | 18 | 22.7 | 26.60 ± 0.53 | 131.00 ± 1.96 | Absent | 39.66 | 0.5 | 0 |
| 2 | CSR-30 | 164.5 | 16 | 21.2 | 31.12 ± 0.57 | 89.3 ± 1.20 | Small | 31.08 | 18.8 | 100 |
| 3 | IC-R28 a | 152 | 14 | 26.8 | 27.32 ± 0.34 | 95.25 ± 0.42 | Small | 31.11 | 0.34 | 97.1 |
| 4 | IC-R32 a | 152.5 | 9 | 25 | 22.62 ± 0.69 | 81.50 ± 0.73 | Small | 33.22 | 0.31 | 91.2 |
| 5 | IC-R38 | 144 | 12 | 22.7 | 26.1 ± 0.58 | 103.75 ± 0.53 | Small | 29.12 | 0.20 | 89 |
| 6 | IC-R42 a | 151.5 | 11 | 25.5 | 26.05 ± 0.26 | 74.75 ± 0.70 | Absent | 36.78 | 0.42 | 89.2 |
| 7 | IC-R58 | 152.5 | 9 | 26 | 29.87 ± 0.52 | 86.75 ± 0.82 | Small | 31.36 | 0.49 | 81.7 |
| 8 | IC-R59 | 151 | 8 | 27.8 | 24.57 ± 0.42 | 78.87 ± 0.52 | Small | 32.89 | 0.25 | 85.8 |
| 9 | IC-R63 | 153.5 | 10 | 26.6 | 26.42 ± 0.32 | 89.00 ± 1.28 | Small | 29.80 | 0.33 | 82.2 |
| 10 | IC-R68 a | 143 | 11 | 27.7 | 30.20 ± 0.40 | 91.25 ± 0.51 | Small | 33.38 | 0.30 | 94.3 |
| 11 | IC-R69 | 141 | 10 | 22.9 | 25.37 ± 0.61 | 87.75 ± 0.85 | Small | 29.36 | 0.46 | 83 |
| 12 | IC-R74 | 154 | 10 | 25.2 | 26.20 ± 0.48 | 105.3 ± 0.51 | Small | 33.63 | 0.29 | 87.4 |
| 13 | IC-R81 | 151 | 9 | 25.7 | 27.30 ± 0.46 | 71.75 ± 0.73 | Small | 31.34 | 0.36 | 82.3 |
| 14 | IC-R90 | 138 | 10 | 24.9 | 26.85 ± 0.37 | 34.00 ± 1.04 | Small | 34.15 | 0.36 | 78.2 |
| 15 | IC-R94 | 130.5 | 9 | 29.8 | 24.97 ± 0.36 | 53.75 ± 0.70 | Absent | 34.21 | 0.31 | 81.8 |
| 16 | IC-R95 | 149.5 | 10 | 24.4 | 32.02 ± 0.20 | 58.75 ± 1.03 | Small | 34.26 | 0.46 | 72.9 |
| 17 | IC-R111 | 142 | 8 | 23.4 | 21.12 ± 0.24 | 69.00 ± 1.03 | Absent | 32.67 | 0.42 | 75.9 |
a Indicates three-gene-pyramided genotypes with maximum RPG.
A significant variation was observed among the pyramided genotypes and parental rice genotypes for grain quality traits (Table 3). Kernel length/breadth ratio after cooking among pyramided BC3F1genotypes varied between 4.2 (IC-R94) to 5.8 (IC-R90), respectively. The elongation ratio for CSR-30 was recorded as 1.93. The elongation ratio among pyramided BC3F1genotypes varied between 1.37 (IC-R38) to 1.64 (IC-R28), respectively. The BC3F1 three-gene-pyramided genotypes except IC-R59 and IC-R94 (ASV-2, high gelatinization, 75–70∘C) showed an ASV of 4 (intermediate gelatinization, 70–74∘C) or 6 (low gelatinization, 55–60∘C). The percent amylose for the pyramided genotypes varied from 21.4% to 35.0%. The four three-gene-pyramided genotypes IC-R28, IC-R68, IC-R32 and IC-R42 were found to have amylose percentages of 24.9%, 24.3%, 25.3% and 22.2%, within an intermediate range similar to that of CSR-30 (23.3%).
Physical grain quality and cooking quality traits of parental and three-gene-pyramided BC3F1 genotypes.
| S. No. | Plant No. | GL (mm) | GB (mm) | KLBC (mm) | KBBC (mm) | KLAC (mm) | KBAC (mm) | KER | ASV | AC (%) | AR |
| 1 | IRBB-60 | 9.72 ± 0.13 | 1.82 ± 0.05 | 7.80 ± 0.26 | 1.89 ± 0.07 | 10.80 ± 0.08 | 2.17 ± 0.02 | 1.38 | 6 | 28.1 | NS |
| 2 | CSR-30 | 8.01 ± 0.09 | 1.80 ± 0.03 | 6.10 ± 0.21 | 1.60 ± 0.04 | 11.77 ± 0.06 | 2.20 ± 0.04 | 1.93 | 4 | 23.3 | SS |
| 3 | IC-R-28 a | 10.91 ± 0.37 | 1.90 ± 0.03 | 7.26 ± 0.07 | 1.66 ± 0.03 | 11.91 ± 0.16 | 2.37 ± 0.08 | 1.64 | 4 | 24.9 | SS |
| 4 | IC-R-32 a | 9.72 ± 0.09 | 1.87 ± 0.03 | 7.14 ± 0.12 | 1.58 ± 0.02 | 11.65 ± 0.11 | 2.23 ± 0.00 | 1.63 | 4 | 25.3 | SS |
| 5 | IC-R-38 | 10.62 ± 0.14 | 1.79 ± 0.06 | 8.44 ± 0.07 | 1.67 ± 0.02 | 11.56 ± 0.03 | 2.12 ± 0.04 | 1.37 | 4 | 26.7 | MS |
| 6 | IC-R-42 a | 10.79 ± 0.21 | 1.85 ± 0.02 | 7.22 ± 0.15 | 1.66 ± 0.02 | 11.02 ± 0.17 | 2.00 ± 0.01 | 1.53 | 4 | 22.2 | SS |
| 7 | IC-R-58 | 10.54 ± 0.15 | 1.87 ± 0.06 | 7.48 ± 0.21 | 1.63 ± 0.04 | 10.88 ± 0.06 | 2.08 ± 0.03 | 1.45 | 4 | 26.6 | MS |
| 8 | IC-R-59 | 10.34 ± 0.15 | 1.90 ± 0.00 | 7.18 ± 0.09 | 1.64 ± 0.03 | 10.59 ± 0.09 | 1.96 ± 0.03 | 1.48 | 2 | 31.7 | SS |
| 9 | IC-R-63 | 9.88 ± 0.22 | 1.86 ± 0.06 | 7.91 ± 0.16 | 1.65 ± 0.02 | 11.63 ± 0.14 | 2.55 ± 0.02 | 1.47 | 6 | 21.4 | SS |
| 10 | IC-R-68 a | 10.02 ± 0.09 | 1.97 ± 0.03 | 7.53 ± 0.05 | 1.67 ± 0.02 | 11.72 ± 0.21 | 2.68 ± 0.02 | 1.56 | 4 | 24.3 | SS |
| 11 | IC-R-69 | 10.49 ± 0.12 | 1.76 ± 0.13 | 7.28 ± 0.05 | 1.64 ± 0.04 | 11.18 ± 0.23 | 2.29 ± 0.04 | 1.54 | 4 | 26.7 | SS |
| 12 | IC-R-74 | 10.91 ± 0.02 | 1.66 ± 0.04 | 7.48 ± 0.03 | 1.62 ± 0.06 | 11.73 ± 0.06 | 2.45 ± 0.06 | 1.57 | 6 | 24.8 | SS |
| 13 | IC-R-81 | 9.91 ± 0.21 | 1.90 ± 0.02 | 7.72 ± 0.09 | 1.65 ± 0.02 | 10.67 ± 0.07 | 1.88 ± 0.06 | 1.38 | 4 | 25.6 | SS |
| 14 | IC-R-90 | 10.05 ± 0.15 | 1.92 ± 0.03 | 7.64 ± 0.17 | 1.64 ± 0.04 | 10.81 ± 0.11 | 1.87 ± 0.06 | 1.42 | 6 | 25.0 | SS |
| 15 | IC-R-94 | 10.43 ± 0.07 | 1.96 ± 0.03 | 8.16 ± 0.12 | 1.75 ± 0.03 | 11.82 ± 0.31 | 2.79 ± 0.09 | 1.45 | 2 | 35.0 | SS |
| 16 | IC-R-95 | 9.77 ± 0.04 | 1.85 ± 0.03 | 7.60 ± 0.14 | 1.62 ± 0.02 | 11.24 ± 0.04 | 2.01 ± 0.04 | 1.48 | 4 | 29.2 | MS |
| 17 | IC-R-111 | 10.22 ± 0.15 | 1.85 ± 0.03 | 7.79 ± 0.03 | 1.67 ± 0.03 | 10.78 ± 0.08 | 2.00 ± 0.00 | 1.39 | 4 | 25.5 | MS |
a Indicates three-gene-pyramided genotypes with maximum RPG.
4 Discussion
BB causes severe yield loss and is the major constraint for Basmati rice production. Several BB resistance genes have been identified and characterized in non-Basmati rice, and incorporated and pyramided through MAS into aromatic rice [14,22,31]. Often the transfer of resistance genes from non-Basmati rice to Basmati rice impairs its grain and cooking quality characteristics. In this study, we report the successful introgression of BB resistance genes into Basmati variety CSR-30 without yield penalty or impairment of its culinary qualities by adopting a molecular breeding approach involving foreground selection along with phenotypic selection for grain and quality characters ([37], [16]).
The three-gene-pyramided BC3F1 genotypes derived in this study from the cross CSR-30 × IRBB-60, were as effective against the virulent Xoo strain as the donor line IRBB-60. The studies suggested that xa5 was most effective, with shorter lesion lengths, followed by Xa21, while lines with xa13 were susceptible to BB. The results indicated that the genes in combination were more effective against the pathogen than a single gene. Evidence suggested that the synergistic action or quantitative complementation between the resistance genes might result in increasing the level of resistance against the Xoo strain [20,24,30].
Molecular-marker-assisted background selection of recombinants was used to determine the parental genome contribution. A total of 131/428 SSR markers produced polymorphism between the parental genotypes IRBB-60 and CSR-30, and were used to estimate the recovery of the recurrent parental genome in the pyramided BC3F1 genotypes. The percentage recovery of RPG in the pyramided BC3F1 genotypes ranged from 72.9% to 97.1%, as revealed by the global statistics using the software Graphical Geno Types (GGT) version 2.0. The low background recovery observed in some genotypes can be due to linkage drag. In the present study, 72.9–97.1% RPG recovery was achieved at the BC3F1 generation, compared to the theoretical expected value of approximately 93.0%. Combining the marker-based foreground selection with the phenotypic selection under the target environment has resulted in more than 90% recovery of the RPG in just three backcrosses, which is similar to what had been observed in most of the studies, where both the foreground and background selections were performed using markers. Therefore, a phenotypic selection combined with marker-assisted background selection can make the breeding protocol economic and effective [24]. Chen et al. [38] reported 98.8% recovery after three backcrosses while selecting for recurrent-parent genome. Neeraja et al. [39] developed the submergence-tolerant rice cultivar Swarna and were able to recover 96.2% RPG in BC3F1 generation. Joseph et al. [22] suggested that the addition of one round of background selection in BC1F1 along with phenotypic selection may greatly increase the efficiency of the breeding program. Most of the three-gene-pyramided BC3F1 genotypes were similar or superior to the recurrent parent CSR-30 for the agronomic traits, grain, and cooking quality traits. The pyramided BC3F1 genotypes were shorter than the recurrent parent CSR-30 and had heavier grain weight, greater than that of the recurrent parent. The genotypes with maximum recovery of RPG, IC-R28, IC-R32, IC-R42, and IC-R68 showed grain yield higher than that of the recurrent parent CSR-30. The recombinants, IC-R42 and IC-R111 also showed the absence of awns as a desirable feature. Aroma, gelatinization temperature (GT) (scored by ASV), and amylose content are the most important characteristics with respect to rice cooking qualities. The three-gene-pyramided BC3F1 genotypes exhibited physical and cooking grain quality characteristics similar to those of CSR-30 (kernel length, kernel breadth, aroma, GT, amylose content). Aroma ranged from mild to strong scented for various recombinants, which may be due to temperature in the grain filling period [40]. All the pyramided genotypes except IC-R59 and IC-R94 (ASV-2, high gelatinization, 75–70∘C) showed an ASV of 4 (intermediate gelatinization, 70–74∘C) or 6 (low gelatinization, 55–60∘C). The cooking time of the rice depends on the coarseness of the grain. The intermediate ASV indicated medium disintegration and is also classified as intermediate GT, which is highly desirable for grain quality [41]. The percent amylose for the pyramided genotypes varied from 21.4% to 35.0%. The four three-gene-pyramided genotypes IC-R28, IC-R68, IC-R32, and IC-R42 had amylose percentage similar to that of CSR-30. Rice with intermediate amylose content (20–25%) has been reported to cook moist and remain soft when cooled, and is widely preferred over rice with high (25–35%) or low amylose contents (10–20%) [42]. The amylose content of the rice influences the glycaemic index (GI) value: rice that is high in amylose has usually a lower GI value [43]. Rice with more amylose has reduced glycaemic responses and lower GI values than a rice variety with less amylose (1–20%) [44]. Thus, rice with intermediate amylose content (20–25%) should have low GI and may be good for consumption by diabetic people. Therefore, all the selected three-gene-pyramided BC3F1 genotypes were found to meet the Basmati grain quality standards.
5 Conclusion
CSR-30 possesses excellent grain, cooking, and eating quality features. Pyramided lines selected from cross CSR-30 IRBB-60 showed grain quality features similar to those of the recurrent parent, CSR-30. In cross CSR-30 × IRBB-60, IRBB-60 as the donor parent for BB resistance facilitated the recovery of backcross-derived plants possessing the desirable grain quality features of CSR-30, despite the donor parent is not a Basmati. Based on agronomic evaluation, BB reaction, percentage recovery of recurrent parent genome, grain quality evaluation and aroma, four genotypes, viz., IC-R28, IC-R68, IC-R32 and IC-R42, were found promising.
Traditional Basmati varieties are highly susceptible to BB disease, for which many effective genes have been tagged and cloned. More resistance genes need to be identified and pyramided together into the elite cultivars to ensure the durability of BB resistance. Most of the major genes have been overcome by new or unrecognized pathogen races. Fortunately, this can be prevented by combining them with other resistance genes. In this study, genes conferring broad spectrum resistance were successfully introgressed through MAS. The pyramided genotypes performing better or equivalently to CSR-30 in grain yield and quality in addition to resistance against BB should be further evaluated at different locations. The pyramided genotypes obtained in our study can be used as genetic resources for BB resistance in breeding programs that will help to achieve better disease management. Marker-assisted selection thus can be successfully utilized to transfer combinations of BB resistance genes into other preferred rice cultivars without compromising yield and quality traits. Advanced basmati breeding lines, which will be derived through MAS and phenotypic selection, will, therefore, be of practical value in providing durable BB resistance in the Basmati growing region and are expected to have a high impact on the yield stability and sustainability of Basmati rice production.


