1. Introduction
Sextonia rubra (Mez.) van der Werff (Lauraceae) is a tropical tree endemic to the Guiana Shield and the Brazilian Amazonia [1]. It is known for its heartwood natural durability and its use as a building material [2]. The natural durability of its wood comes from two main extractives, rubrynolide and rubrenolide, both of which have antifungal and insecticidal activity [3, 4, 5, 6].
Although this species has been known since 1889, its fruit had never been studied in terms of chemical composition. Moreover, no report of any use is described in the literature.
Fruit generally contains specialized metabolites involved in the attraction of pollinators or the deterrence of pathogens/predators, thus involved in their dispersal and defense[7, 8, 9, 10]. It should be noted that the pollinators of S. rubra flowers are still unknown [11]. Specialized fruit metabolites belong to various molecular families [8, 12]. These are of high interest from economic and health points of view due to the antioxidant or antimicrobial activities of fruit essential oils and hydrolates [13, 14].
The characterization of the volatile organic compounds (VOCs) is routinely performed by gas chromatography (GC) coupled to a mass spectrometer (MS) or a flame ionization detector (FID). The data generated can be compared to those present in databases such as the National Institute of Standards and Technology (NIST) or Wiley’s SpectraBase [15]. More recently, we introduced a workflow for GC-EI-MS data analysis that includes the visualization of the spectra collection as a molecular network [16, 17]. This method allows the fast and reliable classification of the EI-MS (electron ionization) data leading to accurate molecule class identification together with home-made databases. We converted the NIST and Wiley spectral libraries in order to make them interpretable by the MetGem software. Headspace-solid-phase microextraction-GC-MS (HS-SPME-GC-MS) [18] and molecular networking analysis were combined to identify the VOCs diversity of S. rubra dried fruit and its hydrolate. The biological activity of the hydrolate against 3 human pathogens, namely Candida albicans, methicillin-resistant Staphylococcus aureus and Tricophyton rubrum, was tested. Finally, its cytotoxic activity was studied with human MRC-5 cell line.
2. Materials and methods
2.1. Plant material
During a sampling campaign in French Guiana in October 2020, a mature tree, belonging to the species Sextonia rubra, was sampled (5°16′846′′ N 52°54′938′′ W, near the Paracou research station, between Kourou and Sinnamary, in a typical terra firme forest area; Permit: ABSCH-CNA-FR-240495-4). Its fruit, probably immature (see Figure S4 in Supplementary Data), was collected and then stored at +4 °C in the field and then at −80 °C pending analysis. Finally, the pieces of fruit were dried at room temperature (20–25 °C) due to their small volume (<2 cm3) and then finely ground in a mortar.
2.2. Hydrodistillation
A weight of 50.13 g of dried fruit was introduced into a flask with a volumetric capacity of 1 L into which 500 mL H2O Milli-Q® was introduced. The flask was connected to a Clevenger apparatus, covered with aluminum foil, and heated to 80–90 °C (see Figure S1 in SI). The coolant for the VOC condensation was a mixture H2O/EtOH (30%/70%, v/v) cooled to 0 °C. The first condensate was obtained after 2 h of heating and collected up to 5 h to obtain a total volume of 18 mL of hydrolate. Hydrodistillation was repeated twice.
2.3. HS-SPME-GC-MS analyses
A volume of 5 mL of hydrolate or 5 g of dried fruit was introduced into a headspace vial (HS), the liquid/gaseous phase equilibration took place for more than 24 h, and then exposed to a conditioned (260 °C for 20 min) solid-phase micro-extraction (SPME) fiber Divinylbenzene Carbon Wide Range Polydimethylsiloxane (DVB/C-WR/PDMS), 80 μm, Agilent Technologies, Waldbronn, Germany) for 5 min without heating. The split ratio was 100:1 for the hydrolate sample and splitless for the dried fruit sample. Following this step, the compounds retained by the fiber were desorbed by introducing the fiber into the GC injection port at 250 °C for 3 min and data acquisition was simultaneous with desorption. The analyses were performed on an Agilent 7890B-GC (Agilent Technologies, Waldbronn, Germany) coupled to an Agilent 7000D triple-Quadrupole-GC (Agilent Technologies, Waldbronn, Germany). Volatile compounds were separated on a HP-5ms column (30 m × 0.25 mm × 0.25 μm, Agilent Technologies, Waldbronn, Germany) subjected to the following temperature gradient: initial temperature +50 °C, ramped at +20 °C⋅min−1 to 150 °C, then ramped at +10 °C⋅min−1 to +210 °C, finally ramped at +20 °C⋅min−1 to +320 °C. The chromatographic conditions were optimized to separate the molecular diversity of each molecular family. Helium was the carrier gas at a flow rate of 1.2 mL⋅min−1. Detection parameters were set as follows: electron energy 70 eV, ion source temperature +150 °C, MS quadrupole temperature +230 °C, mass scanning range m/z 50–500, scanning rate: 3.5 scans⋅s−1.
2.4. Data processing
Chemical diversity analysis of fruit VOCs was performed on MassHunter Unknown Analysis (Agilent, B.09.00), using the coupled spectral libraries from Wiley (2011) and NIST (2017). Default parameters were retained for the analyses with the exception of annotation assignment where only those with a score above 80% were considered. The retention indexes were determined by querying databases. In parallel, molecular networks of the HS-SPME-GC-MS data were created [16]. The raw data in .d were converted to .mzML using the Peak Picking function of MSConvert (v. 3.0.20344), a module of the Proteowizard package [19], the CWT algorithm with “SNR = 0”, and “Peak Spacing = 0”. The .mzML files were then imported into MZmine (v. 2.53) [20]. Only the ions detected above 500 arbitrary unit (a.u.) of intensity were kept in the analyses: “Noise level = 500”. A chromatogram construction step according to the ADAP algorithm was performed with the following parameters: “minimum group size = 3 scans”, “group intensity threshold = 500”, “minimum highest intensity = 500”, “m/z tolerance = 0.3 or 900 ppm”. Then chromatogram deconvolution was performed according to the following parameters: “S/N threshold = 7”, “minimum feature height = 500”, “coefficient area threshold = 50”, “peak duration range = 0.01–0.6 min” and “retention time (RT) wavelet range = 0.01–0.06”. A spectral deconvolution was performed using Multivariate Curve Resolution algorithm with “deconvolution window width = 0.2 min”, “RT tolerance = 0.05” and “minimum number of peaks = 1”. A list of peaks was generated by alignment using the “Join Aligner” function with “m/z tolerance = 0.3 or 900 ppm”, “weight for m/z = 80”, “RT tolerance = 0.1 min” and “Weight for RT = 20”. The data were exported in .mgf and the metadata in .csv.
2.5. Molecular network by MetGem
Molecular networks were created using MetGem software (v. 1.3.6 – 20220401 – Nightly Cl builds: https://github.com/metgem/metgem/releases/tag/nightly). The parameters for the cosine score calculation were: “m/z = 0.3”, “minimum matched peaks = 3” (MMP) and the option “treat as MS1 data” was used. t-SNE representation must be performed with MMP ⩾ 3 for electroionization analyses. The parameters to create the t-SNE representation were: “cosine score(s) above: 0.6”, “number of iterations = 10,000”, “learning rate = 10”, “perplexity = 6” and “early exaggeration = 12”. Annotation was performed by querying the coupled spectral libraries from Wiley (2011) and NIST (2017). The MMP used to match the experimental spectra to those in the databases was 5, although many peaks are present in the spectra obtained by electroionization. This parameter can be modified after an initial analysis according to the molecular families studied. The data in native format were converted by the software Lib2NIST Library Conversion Tool v. 1.0.6.3 into .msp, a format interpretable by MetGem. Then the .msp was modified by replacing the “Names” in “NAMES” and the “Exact Mass: x” in “Exact Mass: 0” so that no correspondence of m/z of parent ions is established between the databases and the data resulting from the analyses in HS-SPME-GC-MS.
2.6. Antimicrobial assays
The hydrolate antimicrobial activity was determined against the following pathogenic strains: Candida albicans ATCC10231, methicillin-resistant Staphylococcus aureus (MRSA) ATCC33591, Tricophyton rubrum SNB-TR1. A negative control without microorganism was performed. Fungal and bacterial cultures are performed with Roswell Park Memorial Institute (RPMI) and Mueller Hinton media respectively. The concentration of essential oil (EO) in the hydrolate was unknown, hence we performed dilutions of the hydrolate in RPMI or Mueller–Hinton culture medium [21, 22]. Eight dilutions were performed in duplicate for each microorganism with the following hydrolate: culture medium ratios: 100:0, 80:20, 60:40, 20:80, 10:90, 5:95, and 2.5:97.5, in other words dilutions to 0%, 20%, 40%, 80%, 90%, 97.5%. Negative controls with sterile H2O Milli-Q® followed the same relative concentrations as the tests. The positive controls were performed using antibiotics solubilized in dimethyl sulfoxide (DMSO, Sigma-Aldrich, Saint-Quentin-Fallavier, France) to obtain the following mass concentrations: vancomycin at 0.64 mg⋅mL−1 against MRSA and fluconazole at 1.28 mg⋅mL−1 against C. albicans and T. rubrum before cascade dilutions. The reading of the activity tests against MRSA was performed after 24 h, 3 days for C. albicans and 5 days for T. rubrum. Each test was performed in duplicate and was repeated twice.
2.7. Cytotoxic assays
The MRC-5 cell line derived from normal lung tissue was obtained from the American Type Culture Collection (Rockville, MD, USA), and was cultured according to the supplier’s instructions. Human MRC-5 cells were grown in DMEM supplemented with 10% fetal calf serum (FCS) and 1% glutamine. The cell line was maintained at 37 °C in a humidified atmosphere containing 5% CO2. Cell viability was determined by a luminescent assay according to the manufacturer’s instructions (Promega, Madison, WI, USA). For IC50 determination, the cells were seeded in 96-well plates (3 × 103 cells/well) containing 100 μL of growth medium. After 24 h, the cells were treated with the hydrolate at 3 different relative concentrations, i.e., 10%, 5% and 1%. Each concentration was obtained from the stock solution. Experiments were performed in triplicate. After 72 h of incubation, 100 μL of CellTiter Glo reagent was added for 15 min before recording the luminescence with the spectrophotometric plate reader PolarStar Omega (BMG LabTech). The dose-response curves were plotted with Graph Prism software, and the IC50 values were calculated from the polynomial curves using the Graph Prism software.
3. Results
3.1. Characterization of the volatile chemical diversity of S. rubra hydrolate and dried fruit by conventional methods
The dried fruit was hydrodistilled to obtain a hydrolate. It smells like cut grass and to a lesser extent like mango. VOCs with a cut grass smell are mainly C6 then C7 and C9 oxygenated molecules such as n-hexanol, n-hexanal, n-heptenal, n-nonanol, n-nonanal, n-nonanone and other saturated or unsaturated alcohols and aldehydes [23, 24]. Among these compounds, n-hexanal, 2-hexenal, heptanal and nonanal were detected but at low relative intensities (<1% for each). Some characteristic molecules of the mango fruit odor are also detected in the S. rubra fruit hydrolate, such as n-hexanal, 2-hexenal, heptanal, α-pinene, β-pinene, limonene, nonanal and α-humulene. However, certain major mango VOCs such as δ-3-carene, terpinolene and myrcene were not detected (see Table 1 and Cuevas-Glory et al. [25]).
Analysis of the main VOCs detected by HS-SPME-GC-MS
| Molecular families | Compound name | Retention index (HP5-ms) | Peak area | |
|---|---|---|---|---|
| Dried fruit (%) | Fruit hydrolate (%) | |||
| Acetate | Phenacylidene diacetate | ND | – | 4.2 |
| Aldehyde | n-Hexanal | 800 | – | 0.9 |
| Aldehyde | 2-Hexenal | 853 | – | 0.3 |
| Aldehyde | Heptanal | 901 | – | 0.4 |
| Aldehyde | Nonanal | 1105 | – | 0.1 |
| Alkane | 1,1-Dimethyl-cyclopentane | ND | – | 0.1 |
| Alkene | Dehydro-ar-ionene | 1355 | 0.3 | – |
| Amino acid | Anthranillic acid | ND | – | 0.2 |
| Ester derivatives | 4-Methylbenzoic acid ethenyl ester | ND | – | 0.4 |
| Indene | 1-Ethyl-trans-3-n-heptyl-2,3-dihydroindene | ND | – | 0.8 |
| Ketone | Sulcatone | 987 | – | 0.5 |
| MH | α-Pinene | 939 | 26.8 | 1.6 |
| MH | Camphene | 955 | 1.9 | – |
| MH | β-Sabinene | 961 | 10.3 | – |
| MH | β-Pinene | 964 | – | 0.6 |
| MH | o-Cymene | 1026 | 0.6 | 0.4 |
| MH | 1,4-Dimethyl-4-vinyl-1-cyclohexene | ND | – | 0.6 |
| MH | Camphene | 954 | – | 0.3 |
| MH | (R)-Limonene | 1030 | 0.6 | – |
| OM | Linalool | 1215 | 0.5 | – |
| OM | Eucalyptol | 1028 | 2.8 | 13.5 |
| OM | α-Phenetylethanol | 1061 | – | 0.4 |
| OM | α-Campholenal | 1125 | – | 0.1 |
| OM | Fenchol | 1126 | – | 0.9 |
| OM | Dehydroborneol | ND | – | 0.9 |
| OM | 4-Acetyl-1-methylcyclohexene | 1131 | – | 0.6 |
| OM | (1R)-(2R)-Nopinone | 1142 | – | 0.7 |
| OM | α-Thujol | ND | – | 0.3 |
| OM | Pinocarvone | 1164 | – | 0.3 |
| OM | Borneol | 1165 | – | 1.8 |
| OM | Terpinen-4-ol | 1178 | – | 0.4 |
| OM | (S)-β-Fenchol | 1112 | – | 0.7 |
| OM | (S)-Myrtenal | 1196 | – | 0.4 |
| OM | Cuminal | 1226 | – | 0.1 |
| 1 SH | Cadalene | 1656 | – | 0.3 |
| SH | α-Cubebene | 1351 | 0.5 | 0.2 |
| SH | Clovene | 1396 | – | 0.2 |
| SH | 7-epi-α-Cadinene | 1490 | 0.4 | – |
| SH | α-Copaene | 1376 | 14.4 | 7.3 |
| SH | β-Copaene | 1430 | 0.4 | 0.2 |
| SH | (R)-Sativene | 1396 | – | 0.1 |
| SH | cis-α-Bergamotene | 1433 | 2.4 | 2.2 |
| SH | α-Santalene | 1424 | 1 | 0.9 |
| SH | β-Caryophyllene | 1427 | 6.3 | 7.4 |
| SH | cis-α-Bergamotene | 1433 | 3.1 | 3 |
| SH | cis-β-Farnesene | 1439 | 2.2 | 2.8 |
| SH | γ-Cadinene | 1449 | – | 0.2 |
| SH | α-Humulene | 1456 | 1.4 | 2.6 |
| SH | β-Gurjenene | 1439 | 0.1 | 0.2 |
| SH | γ-Muurolene | 1477 | 2.1 | 3.7 |
| SH | Cuparene | 1502 | – | 2.6 |
| SH | (S)-Germacrene D | 1503 | 8.9 | 3.7 |
| SH | β-Selinene | 1509 | 0.6 | 1.7 |
| SH | α-Muurolene | 1517 | 2.3 | 4.3 |
| SH | α-Bisabolene | 1522 | 1.7 | 3.4 |
| SH | γ-Cadinene | 1534 | 1.3 | 2.4 |
| SH | δ-Cadinene | 1541 | 2.8 | 5.8 |
| SH | α-Calamenene | 1527 | 2.7 | 7.4 |
| SH | Isoledene | 1373 | – | 0.8 |
| SH | α-Dehydro-ar-himachalene | 1523 | – | 0.2 |
| SH | α-Calacorene | 1543 | 0.3 | 2 |
| SH | α-Corocalene | 1629 | – | 0.2 |
| OS | Guaijol-acetate | 1727 | – | 0.2 |
| OS | γ-Amorphene | 1496 | 1 | – |
| OS | β-Caryophyllene epoxide | 1578 | – | 0.8 |
| OS | Caryophylenol | 1568 | – | 0.3 |
| OS | Isoshyobunone | ND | – | 0.1 |
| OS | (S)-Cubenol | 1636 | – | 0.2 |
| Unidentified | Unidentified | ND | – | 0.1 |
Theoretical retention indices are determined by NIST, Wiley and The Pherobase databases. Relative peak areas were calculated using MassHunter. MH: Monoterpene hydrocarbons, OM: Oxygenated monoterpenes, SH: Sesquiterpene hydrocarbons, OS: Oxygenated sesquiterpenes.
VOCs diversity in dried fruit (Figure 1) is mainly represented by sesquiterpene hydrocarbons (18 detected compounds—69.2%), monoterpene hydrocarbons (4 detected compounds—15.4%), oxygenated monoterpenes (2 compounds detected—7.7%), oxygenated sesquiterpenes (1 compound detected—3.8%) and one alkene (1 compound detected—3.8%). The relative peak areas of the main compounds correspond to α-pinene (26.8%—monoterpene), α-copaene (14.4%—sesquiterpene), β-sabinene (10.3%—monoterpene), (-)—germacrene D (8.4%—sesquiterpene), and β-caryophyllene (6.3%—sesquiterpene) (Table 1). The odors of these compounds are described as grassy and/or woody. VOCs diversity in fruit hydrolate (Figure 1) is mainly composed of sesquiterpene hydrocarbons (25 detected compounds—43.1%), oxygenated monoterpenes (14 detected compounds—24.1%), highly volatile families (10 detected compounds—17.2%), oxygenated sesquiterpenes (5 detected compounds—8.6%) and monoterpene hydrocarbons (4 detected compounds—6.9%). The relative areas of the main compounds correspond to eucalyptol (13.5%—monoterpene), α-calamenene (7.4%—sesquiterpene), β-caryophyllene (7.4%—sesquiterpene), α-copaene (7.3%—sesquiterpene) and δ-cadinene (5.8%—sesquiterpene) (Table 1). The odors of these compounds are described as grassy and/or woody.
Chromatograms of the VOCs of dried fruit (upper part) and fruit hydrolate (lower part) by HS-SPME-GC-MS. The chromatogram is colored according to the different molecular families detected and annotated.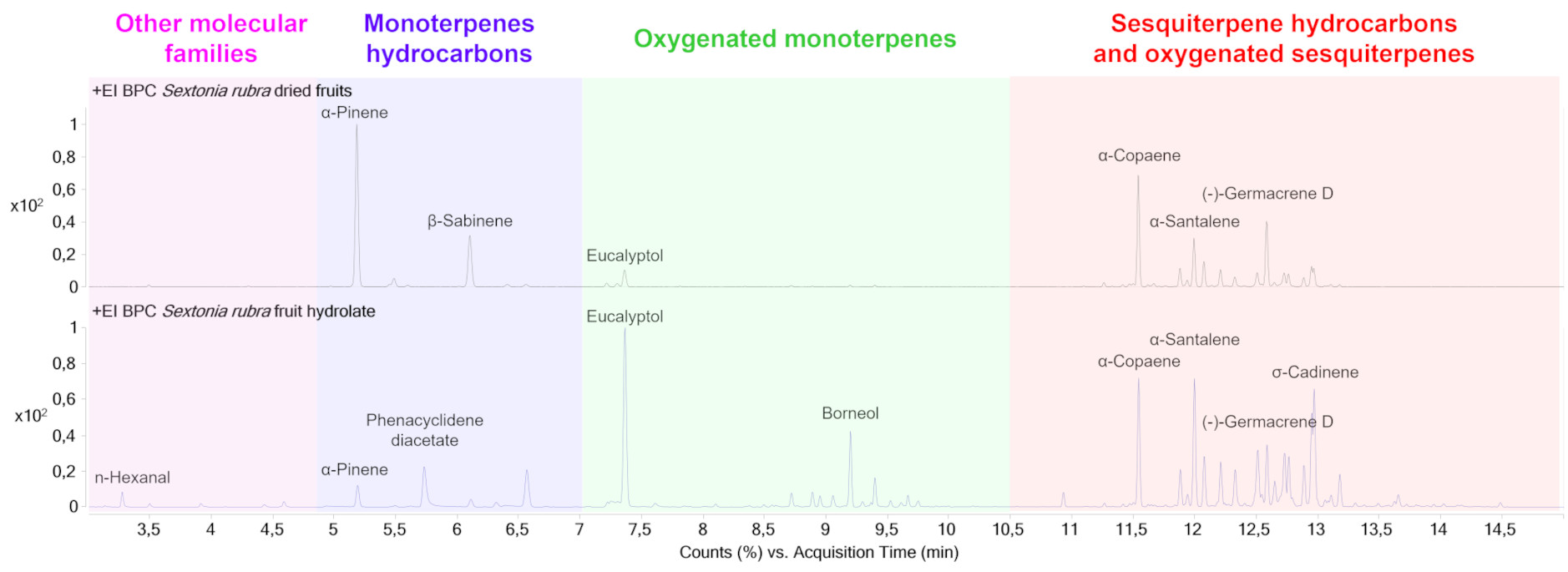
If we compare the sums of the peak relative areas corresponding to the different molecular families between the two samples analyzed, the hydrodistillation process seems to favor the extraction of small highly volatile molecules (+13.4%), oxygenated monoterpenes (+16.4%) and oxygenated sesquiterpenes (+4.8%). On the other hand, the relative abundance of monoterpene hydrocarbons and sesquiterpene hydrocarbons decreased by 8.5% and 26.1%, respectively, in comparison with the dried fruit (see Table S1 in Supplementary Data).
3.2. Characterization of the chemical diversity of S. rubra hydrolate and dried fruit by molecular networks
The resulting data can also be used to create molecular networks by pre-processing the data with MZmine v. 2.53 and MetGem v. 1.4.1 [17, 20]. Molecular networks allow comparison of chemical diversity between different complex samples. Their principle is based on the assumption that structurally close molecules share similar fragmentation patterns. Thus, it is possible to visualize clusters grouping structurally similar molecules and to annotate them by correspondence with spectral banks [26]. Two tools, among others, are currently available for generating molecular networks from mass spectrometry data: GNPS and MetGem [17, 27]. In contrast to GNPS, MetGem is a software package offering the user a certain modularity, i.e. real-time parameter modification, generation of several molecular networks, and the ability to generate a t-SNE representation. This algorithm represents the chemical diversity of one or more samples in a 2-dimensional space, where the distances between nodes are the corollary of the spectral similarity between the studied molecules. This representation can be used to determine whether there are structural similarities between different clusters of the same molecular network [17, 28]. MetGem is able to reprocess MS1 data from both MALDI-ToF and GC-MS analyses [16, 29]. However, GC-MS databases are not open-source, e.g., NIST, Wiley … It is however possible to query them through MetGem after their conversion into a dedicated format.
Figure 2 is a portion of the t-SNE representation of the ions detected in HS-SPME-GC-MS of the dried fruit and hydrolate. Three clusters are distinguishable, with cluster A grouping monoterpene hydrocarbons. Nodes in this cluster share the following fragments m/z 93.1, 91.1, 79.1, 77.0. Cluster B includes oxygenated monoterpenes and monooxygenated molecules with a number of carbon atoms lower than 10 (nC < 10). The nodes constituting cluster B, and being annotated as oxygenated monoterpenes, share the following common fragments m/z 93.1, 81.1, 79.1, 69.1, 67.0. However, common higher molecular weight fragments are shared, partially, between oxygenated monoterpenes and oxygenated molecules with nC < 10. Cluster C groups sesquiterpene hydrocarbons and oxygenated sesquiterpenes. The nodes corresponding to sesquiterpene hydrocarbons share the following common fragments m/z 204.2, 189.2, 161.2, 105.1, 93.1, 91.1 and 79.1. The nodes corresponding to aldehydes, ketones, esters are scattered in the network. Moreover, polysiloxane impurities were detected and annotated but are not represented in Figure 2 (see Figure S1 for more information). These compounds probably come from the SPME fiber used which is composed of polydimethylsiloxane (DVB/C-WR/PDMS).
t-SNE representation of some of the chemical diversity of the VOCs in dried fruit and hydrolate. Cluster A contains monoterpene hydrocarbons, cluster B contains oxygenated monoterpenes and cluster C contains sesquiterpenoids, grouping sesquiterpene hydrocarbons and oxygenated sesquiterpenes. The color code shows the main molecular families. The green nodes are the ions detected in the hydrolate and the red nodes correspond to the ions detected in the dried fruit. The total molecular network contains 1088 nodes, 482 of which are shown in this figure, i.e., those corresponding to VOC. 199 nodes are singletons and 407 correspond to polysiloxane derivatives (shown in Figure S1). Pie-charts correspond to the peak areas of each detected ion using MZmine. Node size is defined by the maximum area for a peak corresponding to an ion detected in both samples.
The clustering performed by MetGem according to the fragmentation patterns of the molecules is specific to each molecular family, i.e. monoterpene hydrocarbons, oxygenated monoterpenes and sesquiterpenoids are distinguished (Figure 2). On the other hand, compounds belonging to highly volatile families as well as oxygenated molecules with nC < 10 are dispersed in the network (Figure 2). The MetGem annotations are consistent with those performed by the MassHunter Unknown Analysis (Agilent, B.09.00) algorithm. This observation is only valid for ions with a high relative intensity (relative area > 0.2%). Indeed, when the intensity is below this threshold, the cosine scores decrease (see Figure S3 in Supplementary Data). This results in uncertain matches with the spectral banks, so it remains and will remain necessary to check the data manually. Moreover, this methodology was not able to annotate the ion detected at Retention Time (RT) = 13.73 min (Table 1). The use of MetGem therefore complements the MassHunter software, as it facilitates the visualization of data obtained from GC-MS analyses. Indeed, it is possible to apply additional filters to this visualization (retention time, common fragments …). In addition, a large number of samples can be analyzed simultaneously to determine whether they share a similar chemical diversity. However, many nodes are annotated as contaminants, in particular polysiloxanes, but belong to clusters not shown in Figure 2 (Figure S1), thereby not interfering with data analysis since the respective surface area of each of these compounds was not taken into account in the analysis.
The exploration and annotation of the chemical diversity of VOC is now possible through the use of numerous libraries. It is possible to annotate quickly and with high confidence the identity of the ions present in a molecular network without using retention indices [27, 30]. This methodology has already been successfully used to search for compounds with larvicidal activity against Aedes aegypti [31].
3.3. Comparison of VOCs composition between different organs of S. rubra
To our knowledge, this is the first time that the volatile chemical composition of the Sextonia rubra fruit has been studied. On the other hand, a previous study carried out on essential oils (EO) of S. rubra leaves led to the identification of 44.6% monoterpenes, 3.2% oxygenated monoterpenes and 47.7% sesquiterpenoids. The EO of branches was made up of 1.2% monoterpenes, 66.8% sesquiterpenes and 26.7% sesquiterpenoids (relative areas) [32]. The monoterpenes common to the hydrolate, dried fruit, branches and leaves of S. rubra are α-pinene and camphene [32]. Sesquiterpenes common to the hydrolate, dried fruit, branches and leaves are α-copaene, β-caryophyllene, α-muurolene, δ-cadinene and α-calamenene [32]. In general, the molecular family most represented in the EO or hydrolates of the different tissues of S. rubra corresponds mainly to the sesquiterpenes with the exception of leaves. The relative VOC compositions are similar between fruit and leaves but differ widely with that of the branches. Alcântara et al. obtained extraction yields of EO from S. rubra fresh leaves and branches of 0.14% and 0.01%, respectively. The hydrodistillation carried out on S. rubra dried fruit resulted in a hydrolate with an extraction yield of 36%.
3.4. VOCs in S. rubra fruit: a perspective
Based on the original composition of the fruit hydrolate, we determined its relative minimum inhibitory concentrations (RMIC) against three human pathogens, i.e. Candida albicans, MRSA and Tricophyton rubrum (Table S2). The concentration of EO in the hydrolate was unknown, hence, we performed dilutions of the hydrolate in RPMI or Mueller–Hinton culture medium. It has been demonstrated that EO rich in sesquiterpenes can exhibit moderate to high antifungal activities [33, 34, 35, 36]. The growth of C. albicans and MRSA was not altered in the presence of the hydrolate, regardless of the concentration used. On the other hand, when T. rubrum was exposed to relative concentrations greater than or equal to 5%, its growth was inhibited. This observation could be due to the high proportion of α-copaene in the hydrolate. Indeed, Houël et al. had determined a MIC of α-copaene at 4 μg⋅mL−1 against T. rubrum [34]. Moreover, cuminal, although detected in low proportion (Table 1), is responsible for a high activity against dermatophyte such as T. rubrum [37]. In view of the mechanism of action of terpenes, the activity of the hydrolate against T. rubrum could be similar towards filamentous fungi sharing a cell membrane close to that of T. rubrum [38, 39, 40]. Despite the anti-fungal activity observed against T. rubrum, the hydrolate did not show any cytotoxic activity against the MRC-5 cell line at relative concentrations of 10%, 5%, 1% (Table S3).
By comparing the current scientific literature with our study, the EOs composition of S. rubra (obtained from: bark, branches, fruit, heartwood, leaves, seeds, stem and twigs) is dominated by β-caryophyllene and eucalyptol which are detected in the EO and hydrolate prepared from different organs of S. rubra [32, 41]. Lauraceae’s EO are characterized by a high abundance in hydrocarbons and oxygenated sesquiterpenoids, which is consistent with the relative intensities of the molecular families detected in the fruit hydrolate (Table S1). In the Lauraceae family, the main VOCs detected are β-caryophyllene, germacrene D, bicyclogermacrene, caryophyllene oxide, α-bisabolol and byclogermacrenal. In the two samples analyzed, β-caryophyllene and germacrene D are detected with relative intensities greater than 5% [42].
Moreover, our study raises new questions on the ecological functions of sesquiterpenes detected in S. rubra fruit. Interestingly some compounds exhibit insecticidal activity, such as β-caryophyllene (>5%) and α-humulene (>1%), against Aedes aegypti, and describing the spectrum of insecticidal activity of these compounds would be of interest [43, 44]. The same applies to α-humulene (>1%) against Sitophilus zeamais and Tribolium castaneum [45]. Additionally, α-copaene (>5%) and sulcatone (0.5%) could be an attractant for some arthropods [30, 46, 47]. β-caryophyllene (>5%) has also been identified for its role as an attractant for Elsholtzia rugulosa’s pollinators and for its ability to inhibit the growth of pathogenic bacteria [48, 49]. Eucalyptol, a major component of the VOCs of this fruit (2.8% in dried fruit, 13.5% in hydrolate), has been reported to have anti-fungal activity against phytopathogenic fungi Aspergillus flavus, Botrytis fabae and Uromyces fabae [50, 51]. Although present in small quantities in the hydrolate (0.2%), anthranillic acid has also been reported to have antifungal activity against Botrytis cinerea [52]. Eventually, some terpenes are known to promote plant growth such as β-caryophyllene (>5%) and α-pinene (26.8% in dried fruit, 1.6% in hydrolate) [46, 53]. In view of the maturity of the fruit in comparison with the literature, it is possible that it emits these two compounds in order to promote their maturation [1, 54]. However, these suppositions require a better understanding of the biology of S. rubra, including the identification of its pollinators and pathogens to elucidate the role of these compounds in the ripening and protection of its fruit.
4. Conclusions
In this work we identified 98.5% of the number of peaks corresponding to VOCs emanating from the S. rubra dried fruit and hydrolate by HS-SPME-GC-MS using the NIST and Wiley databases. The analysis performed on MassHunter Unknown Analysis served as a guide to verify the annotation results obtained by MetGem. The use of MetGem proves to be a complementary methodology to the use of MassHunter, as numerous filters can be used when processing data from a large number of samples. In addition, MetGem can now be used to query databases specific to GC-MS analyses. Finally, the hydrolate of this fruit has a relative antifungal activity of 5% against T. rubrum. In general, this study presents a protocol to exploit data from GC-MS analyses on MetGem and query spectral banks such as NIST and Wiley participating in the characterization of the chemical composition of the Amazonian fruit diversity. Particular attention should be paid to sequiterpenes in the future to characterize their ecological functions following their synthesis by trees. This work is part of a series of studies designed to characterize the chemical defenses of S. rubra.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.
Acknowledgments
We thank the CIRAD team, Saint-Omer Cazal, Michel Baisie, Soepe Koese, Alpha, and the INRAE tree climber Jocelyn Cazal for the pruning and felling of the studied tree.




 CC-BY 4.0
CC-BY 4.0