1. Introduction
Seaweeds are classified into three phylogenetically independent groups: the red algae (Rhodophyta), the green algae (Chlorophyta), and the brown algae (Phaeophyceae) [1]. Within the class Phaeophyceae, the order Fucales represents nine families and contains more than 500 described species, including the Fucaceae. The family Fucaceae comprises six genera: Ascophyllum, Silvetia, Pelvetia, Hesperophycus, Pelvetiopsis and Fucus [2] (Figure 1).
Scheme showing the phylogenetic positions of Fucaceae genera and picturing representative species from the main orders (B) among the brown algal lineage (A).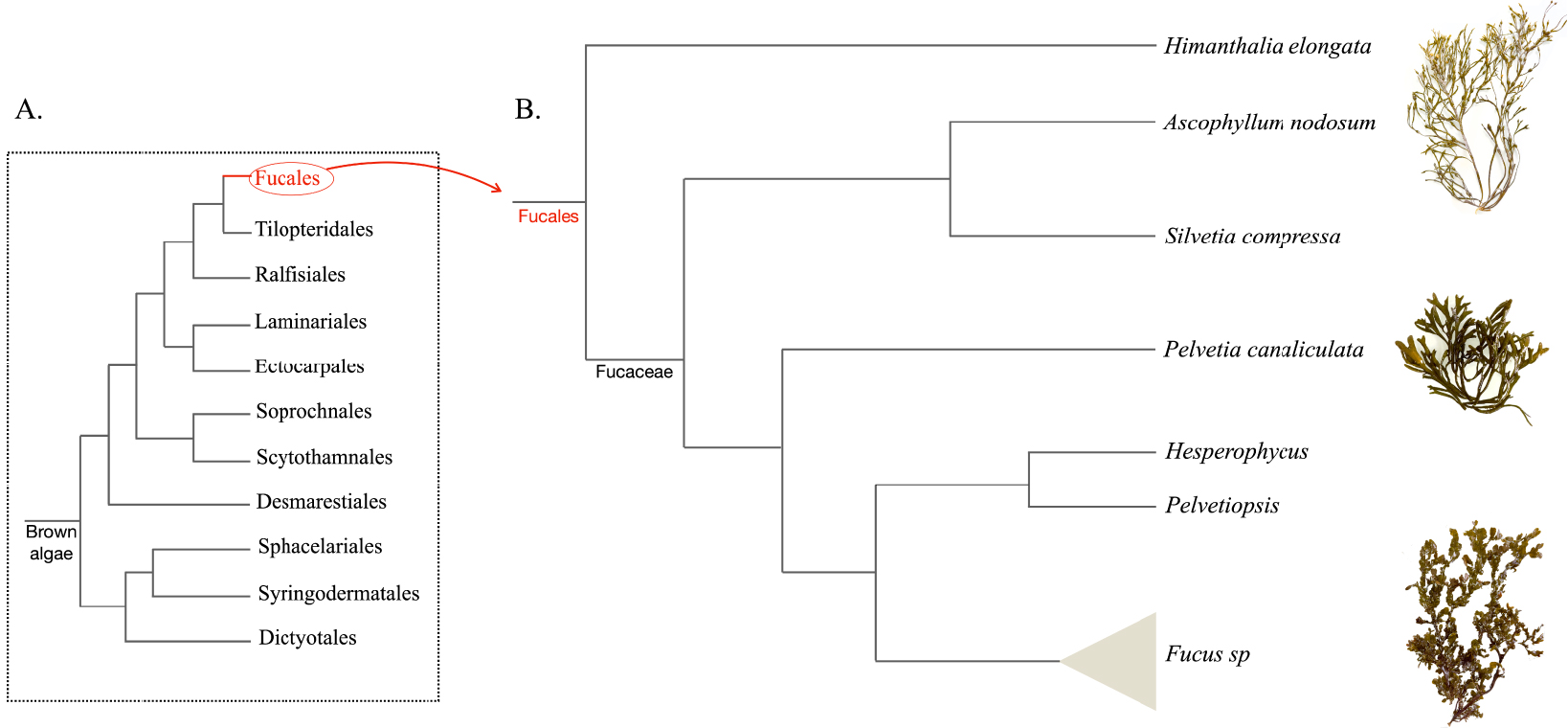
The life cycle of the Fucaceae is monogenetic and heteromorphic with both diecious and monoecious species (Figure 2).
Examples of life cycles in Fucales of dioecious (A) and monoecious (B) species.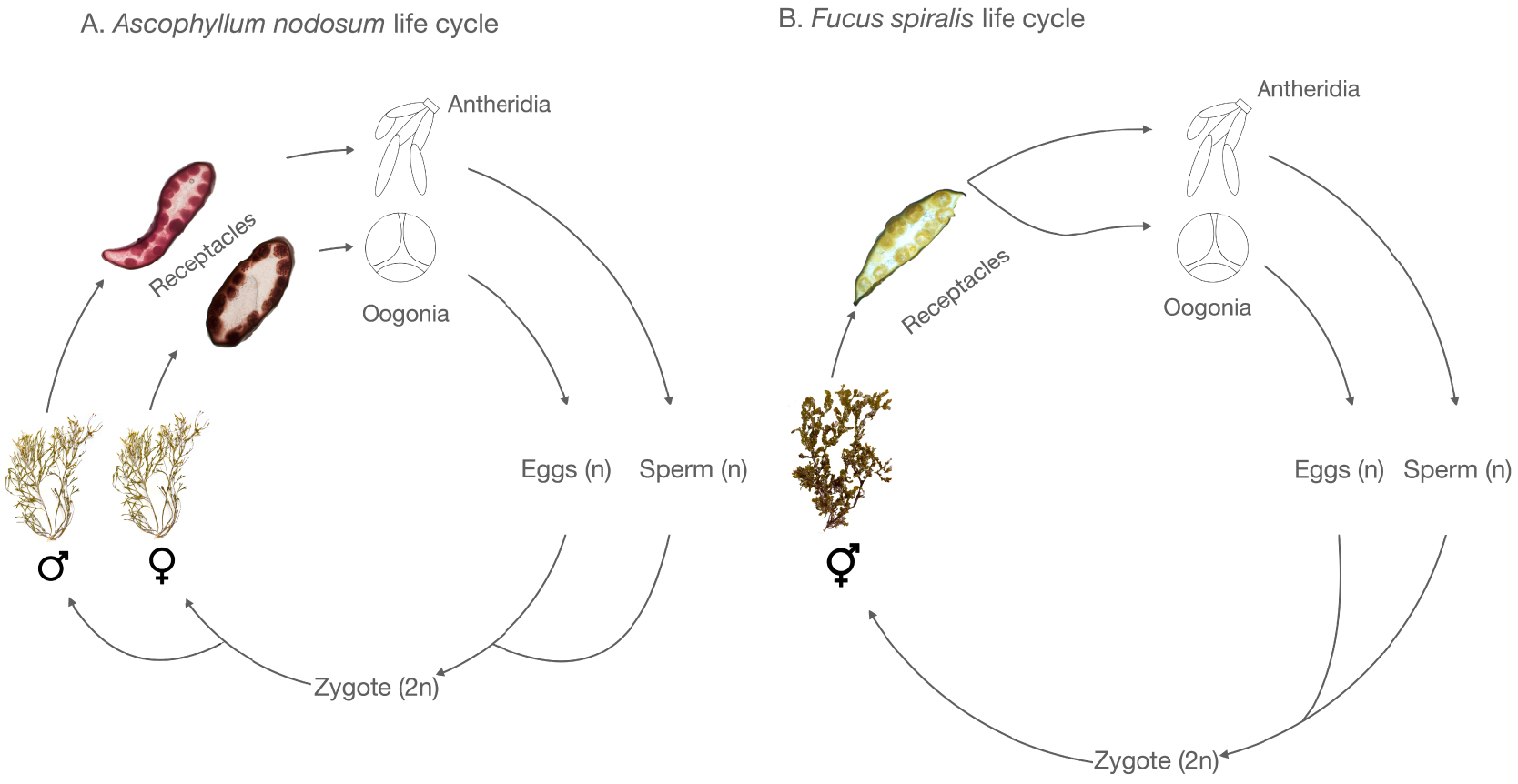
Species of Fucaceae thrive mainly in the Northern Hemisphere in foreshore rocky substrates exposed to the tides, known as the intertidal zone (Table 1). Pelvetia is found in the North-East Atlantic Ocean, the Arctic Ocean, and the North Sea. Ascophyllum and Fucus are mainly present in the North Atlantic Ocean, while Pelvetiopsis, Silvetia, Fucus, and Hesperophycus are found in the North Pacific (Table 1). The changing physicochemical factors of the intertidal zone, such as salinity, solar radiation, tidal range, wave exposure, and temperature influence the geographic distribution of macroalgae. These factors also generate a local distribution pattern such as the vertical zonation. Pelvetia canaliculata is usually found in the extreme upper intertidal zone, followed by Fucus spiralis and Fucus guiryi. In the mid-intertidal zone, Ascophyllum nodosum prefers sheltered rocky shores whereas Fucus vesiculosus occurs on exposed rocks. In the lower intertidal zone, Fucus serratus is commonly observed growing on sheltered rocks [3, 4].
Distribution of the six genera from the Fucaceae family
| Genus | Geographic region | Zone | References |
|---|---|---|---|
| Ascophyllum | North Atlantic Ocean | Sheltered intertidal | [5, 6, 7] |
| Fucus | North Pacific, North Atlantic | Mid-tidal to subtidal | [8, 9, 10] |
| Pelvetia | North-East Atlantic, Arctic Ocean, North Sea | Sheltered extreme upper intertidal | [6, 11] |
| Pelvetiopsis | North Pacific, from South to North Canada | Upper intertidal | [12] |
| Silvetia | North Pacific, West coast of North America, Coastlines of Japan, China and Korea | Mid-tidal | [6, 12] |
| Hesperophycus | North Pacific, from Santa Cruz, California to Islas San Benito, Baja California | Upper intertidal | [13] |
Some members of the Fucaceae are considered ecosystem engineers [6, 9, 10] as they form diverse three-dimensional habitats that serve as refuge for a wide range of micro- to macroscopic living organisms [14, 15]. Lastly, the Fucaceae are important for the marine food web, and act as a source of food for a diverse array of living organisms such as birds, mammals, and fishes [16].
Beyond their ecological functions, the Fucaceae have been harvested by humans for centuries, serving not only as sustenance for livestock and humans, but also finding application in agriculture and as traditional medicinal remedies [12]. Regarding the exploitation of Fucaceae, to our knowledge there are no data available globally or at the European level for this specific family. However, at least in some regions, such as Brittany in France, A. nodosum and Fucus spp. are the main intertidal seaweed species exploited, accounting for 80% of the total amount of seaweed (6436 tons fresh weight) harvested directly on the shore in 2021 by professionals [17, 18]. The strong local exploitation of these two species of Fucaceae is consistent with their distribution in the North Atlantic Ocean (Table 1), as France is the second largest producer after Norway in Europe. Globally, the majority of these uses are based on the alga itself or its extracts. However, the biosynthetic origin of the bioactive molecules within these extracts is not yet fully known. Indeed, some of the compounds present in these seaweed extracts may be derived from the seaweed itself or from its microbiota (i.e., bacteria, archaea, viruses, fungi and other eukaryotes found in the biofilm forming on the host surface but also inside the host tissues).
The aim of this systematic review is to synthetize the current knowledge on aspects of the biology and chemistry of the Fucaceae and their associated microbiota. It will examine the chemical interactions within the Fucaceae holobiont, i.e., the functional entity resulting from the association of a host and its microbiota [19, 20, 21], and will lead to a better understanding of the ecological roles assumed by its different partners. This fundamental knowledge on the chemical ecology of Fucaceae, i.e., chemically mediated interactions between the Fucaceae and associated organisms, will also offer new perspectives on the application and valorization of the Fucaceae holobiont. For instance, it could reveal whether some microorganisms are necessary for the seaweed to produce specific compounds of interest. It also helps us to understand to what extent and how the environmental parameters affect the biosynthesis of compounds within the holobiont in Nature. In the case of molecules produced by a microorganism or a community of microorganisms itself, it would be possible to consider a year-round production of the compound of interest in a bioreactor, which would contribute to the sustainable exploitation of the chemical diversity of Fucaceae holobionts.
We will first review the applications of the Fucaceae (Section 2) focusing on the European market, followed by reviewing the available information regarding the algal microbiota and its influence on the host, such as how fitness and the production of compounds of interest are impacted (Section 3). Lastly, we will focus on the compounds produced by the associated microbiota and discuss their potential role in the biology and ecology of the Fucaceae (Section 4).
2. Overview of the applications of Fucaceae
In Europe, the leading markets for seaweed-derived products are the production of plant biostimulants for agriculture and the phycocolloid industry. Thus, the main commercially exploited species are brown seaweeds (more than 99%), such as Alaria esculenta, Laminaria hyperborea, Laminaria digitata, Saccharina latissima, Undaria pinnatifida, and A. nodosum, which belongs to Fucaceae [22]. Regardless of the field of application, it is important to remember that the chemical composition of a seaweed species and its extracts is affected by, among others, the season and location of harvesting [23, 24, 25]. These factors must be considered since they likely influence the biological activities of the commercial products being manufactured. A general overview of the current uses of the Fucaceae and Fucaceae-derived products is provided in the following subsections according to the type of application. In addition, promising results obtained at a laboratory scale are also presented, suggesting other potential applications for Fucaceae in the coming years.
2.1. Food ingredients and additives
In some regions of the world, the consumption of seaweed as food has been, for many centuries, linked mostly to times of hardship, scarcity, and famine. For instance, during the Irish Great Famine of 1845–1847, seaweed became the ‘last resort’ for the starving population, and the Fucaceae P. canaliculata was one of the seaweeds used as a food source [26]. Nowadays, seaweeds are considered valuable sources of food ingredients due to their content in minerals, soluble and insoluble dietary fibers, proteins with well-balanced essential amino acids, and fatty acids, among others. For instance, Fucus spp. are good sources of Na, K, Ca, Mg, Fe, Cu, Zn, Mn, and I [27, 28]. In Europe, more than 150 species of algae (i.e., microalgae, cyanobacteria, and seaweeds) are consumed but only 30 species are approved by the Novel Food regulation (EU) 2015/2283 for human consumption. Among these 30 species, brown seaweeds are well represented, and species belonging to Fucaceae, A. nodosum, F. serratus, F. spiralis, and F. vesiculosus are the ones approved by the Novel Food regulation [22]. Several studies report the use of F. vesiculosus and to a lesser extent of A. nodosum as valuable food ingredients or food additives, mostly as a source of phlorotannins and antioxidants, which prevent food spoilage resulting from oxidative deterioration. These two families of compounds have already been incorporated in many food types, as powders in bread or more often as an aqueous or ethanolic extract in fish, fish-based products, and dairy products. However, the incorporation of such seaweed-based ingredients frequently compromises the organoleptic profile of the final products [29]. Finally, the main food additives based on brown seaweed are alginates and alginic acids (E400), used as gelling and thickening agents in many food products. European seaweeds contribute to 34% of the world’s alginate and alginic acids supply, and, although it likely only makes up a small proportion of the overall production (precise data are not available) in comparison with other brown seaweeds (e.g., Laminaria spp.), A. nodosum is one of the species processed by this industry. Overall, Europe is the top food and pharma-grade alginate producer in the world [5, 22, 29].
2.2. Human health and well-being
The Fucaceae also have many benefits for human health and well-being, such as specific phenolic compounds (phlorotannins), carotenoids (fucoxanthin), polysaccharides and iodine [2, 12, 27]. According to recent studies, A. nodosum, F. vesiculosus, and Silvetia compressa extracts may help manage and prevent metabolic syndrome and related disorders, which could be in part attributed to prebiotic activities [30, 31]. Phlorotannins represent a unique and diverse class of phenolic compounds exclusive to brown algae and are abundant in the Fucaceae [2]. Phlorotannin contents can reach up to 14% and 12% dry weight in A. nodosum and Fucus spp., respectively [12]. It has been demonstrated that phlorotannins possess numerous health benefits such as antioxidant effects, antidiabetic properties, anti-inflammatory effects, and antitumor properties [12]. Lopes et al. have demonstrated that phlorotannin-rich extracts from Fucus species were able to inhibit carbohydrate-metabolizing enzymes as well as the xanthine oxidase, an enzyme usually overexpressed in patients with diabetes. These results are encouraging for the incorporation of phlorotannin-rich extracts in nutraceuticals or pharmaceuticals for glycemic control and diabetes-related vascular disorders [32]. Brown seaweeds also contain a range of polysaccharides (alginate and alginic acids, fucose-containing sulphated polysaccharides (i.e., fucoidans and ascophyllan), laminaran, or laminarin), but none of them are specific to the Fucaceae. In fact, alginate is the predominant polysaccharide component in the cell walls and intercellular matrix of all species of brown seaweeds. Fucose-containing sulphated polysaccharides, such as fucoidans, are also present in many other brown seaweeds, but unlike alginate, which has a well-defined structure, the chemical structure and composition of fucoidans depend to a large extent on the different species in which they are found. For instance, the structure of fucoidans from two Fucaceae species, A. nodosum and F. vesiculosus, is different, illustrating the huge structural variety of brown seaweed polysaccharides [33, 34, 35]. A wide range of beneficial activities are associated to alginates, fucoidans, and laminarans, such as antioxidant, antimicrobial, antitumor, anticoagulant, prebiotic, immunostimulatory, and antidiabetic properties [36]. They are also used for biomedical applications in biocompatible and biodegradable materials, such as for wound-healing films (e.g., ALGICELL™, SeaSorb®), tissue engineering, and drug delivery applications [37, 38].
2.3. Cosmetics
Brown seaweeds, including the Fucaceae, produce a myriad of bioactive compounds of interest that are used in cosmetic products as well as in thalassotherapy, e.g., Fucus spp. baths [39, 40]. Over the last few decades, the “marine cosmetics” industry has developed considerably, with many companies exploiting the wide range of properties of seaweed, such as Fucus spp. and A. nodosum for “phycocosmetics” [41]. They produce and provide ingredients for the cosmetics industry or develop their own commercial products (e.g., NutraMara, Rí Na Mara, Maiiro, Nuwen, Lessonia, Agrimer cosmetic, Thalion). Alginate is used for its various rheological properties (gelling agents, thickeners, emulsifiers) as well as for its bioactivities (antioxidant, anticellulite, anti-inflammatory, antiphotoaging and moisturizer), which are also described for fucoidans and laminarans [40]. The phenolic compounds of Fucaceae (i.e., the aforementioned phlorotannins) are incorporated into cosmetics as antioxidant, anti-aging/antiwrinkling, anti-allergic, whitening, or UV protective agents. For example, a recent study demonstrated higher anti-aging activities of A. nodosum and F. serratus fractions enriched in phenolic compounds compared to the anti-aging activity of the tea flavonol epigallocatechin gallate, which is a powerful antioxidant already marketed for its anti-aging properties [42]. Another example is a phlorotannin-enriched fraction from F. spiralis, which exhibited a higher antioxidant ability (DPPH assay) in comparison with the synthetic standard Butylated Hydroxytoluene (BHT). This extract also exhibited a strong inhibition of collagenase and elastase making F. spiralis a promising species for dermatological applications [43]. Finally, over the past ten years, the safety of synthetic ingredients in cosmetic products has been questioned. For instance, some synthetic preservatives such as parabens are controversial. They are suspected to cause skin sensitization, allergic dermatitis, as well as estrogenic and carcinogenic effects (breast cancer and malignant melanoma) [44]. Due to their antifungal and antibacterial activities, phenolic compounds of Fucaceae are promising alternatives as natural cosmetics preservatives [40, 45].
More recently, seaweeds have been gaining attention as ingredients in “cosmeceuticals”, which are cosmetic products claiming to have medicinal or drug-like benefits [46]. Indeed, most of the seaweed components and properties mentioned above for human health are also valuable for the cosmetic industry.
2.4. Agriculture
Ascophyllum nodosum and to a lesser extent species belonging to Fucus have been used for centuries by farmers from coastal regions of the northern hemisphere to fertilize agricultural land. These species are recognized for their rich macro- and micro-nutrient content, essential for plant growth, making them biofertilizers of choice. In the last few decades, they have also been harvested to produce tailored extracts used as biostimulants for plant growth and increased tolerance to abiotic stresses. These properties have been linked to their rich content in minerals (e.g., potassium), plant hormones (e.g., auxins, cytokinins, and abscisic acid), osmotic regulators (e.g., glycine betaines), phlorotannins, bioactive carbohydrates (e.g., alginates, oligosaccharides) [5, 47, 48, 49]. For instance, biostimulants made of A. nodosum extracts demonstrated significant benefits on wheat, maize, tomato, sweet pepper, and grapevine crops [50, 51, 52, 53, 54, 55]. Different companies around the world, such as Acadian Seaplants Ltd., Maxicrop Ltd., and Timac Agro have commercialized products derived from A. nodosum extracts [47]. The plant biostimulant market is continuously growing and, according to some projections, the European demand for seaweed-based biostimulants is projected to be worth up to €1.8 billion by 2030 [48, 56]. Recent studies have also demonstrated that A. nodosum extracts could potentially stimulate plant defenses against biotic stresses and be used as an alternative to phytosanitary products to fight plant pathogens like mildew [49, 57]. These benefits also apply to seaweed cultivation, which just like vascular plants are subjected to diseases and epiphyte infestation. Notably, the use of commercial biostimulants based on A. nodosum extracts is being explored for enhanced growth rate, and mitigation of epiphyte and endophyte infestations during the cultivation of red seaweeds such as Kappaphycus sp. [58, 59, 60]. However, the benefits of such A. nodosum-based biostimulants need to be further studied as the active compounds might be dependent on environmental conditions, species and life stage [58, 61].
Brown seaweeds, in particular A. nodosum, are also widely used for livestock feed with positive effects on growth performance, stress and pathogen resistance, and meat quality of the livestock [62]. The rich phenolic compounds in A. nodosum are one of the main factors promoting feed digestibility and animal health in general. The A. nodosum-based Tasco® product from Acadian Seaplants is one example of this [45]. Such effects have been demonstrated in pigs [63, 64] as well as poultry [65], making animal feed a most promising short-term market for seaweed use in Europe as it is projected to reach up to €2.2 billion by 2030 [56]. Finally, the aquaculture industry is also an important market as it generates up to 50% of the world’s fish production for human consumption. Here too, extracts of Fucaceae, especially those from A. nodosum, are promising for the production of natural feed additive owing to their antibacterial, antifungal, antibiotic, antioxidant activities and phenolic compounds which could also be used as a natural cross-linker for aquaculture food delivery systems, for examples as fish oil microencapsulation [66, 67, 68].
2.5. Biomass availability and sustainability
To sustain the growing demand for Fucaceae biomass of these different markets, its source must be abundant and renewable. Countries exploiting wild seaweed off the Atlantic coast have different management practices according to the species being harvested [5]. Even though the harvesting of A. nodosum has been considered to be economically sustainable for centuries in Northern Europe, there are recent concerns about its environmental impacts. A recent study recommended avoiding harvesting A. nodosum from the wild in Portugal because of the vulnerability and ecological importance of this southernmost population in Europe [69]. In Eastern Canada, where the commercial exploitation of A. nodosum began in the late 1950s, studies evaluating the impact of harvesting and management over 25 years concluded that A. nodosum harvesting is sustainable in this area, however it needs to be closely monitored as environmental conditions are changing [7, 70, 71]. In Alaska, the harvesting of Fucus distichus is also considered sustainable due to a tailored management plan which takes into account the timing of reproduction, the available biomass, and regrowth [72]. In Europe, the harvesting of Fucus spp. from wild populations is permitted in some countries (i.e., Ireland, France) and prohibited in others (i.e., Germany) [73]. In the future, regardless of the Fucaceae species, local harvesting and monitoring practices should be adapted to ensure sustainable management of wild resources to avoid overharvesting at a global scale [7, 69, 70, 71] and it may become necessary to investigate different strategies for Fucaceae cultivation in coastal and/or offshore waters [5, 73].
3. Seaweed and their microbiota, a complex holobiont
We believe that the microbiota of Fucaceae may contribute to finding suitable responses to a sustainable commercial exploitation of these seaweed species. As reviewed below, the Fucaceae microbiota is complex and may contribute significantly to the health of the host alga but also its chemical composition including the production of bioactive compounds. A better understanding of the Fucaceae microbiota could lead to the development of alternative production methods of compounds of interest, for instance by directly synthesizing and/or producing the compounds of interest from microbial culture.
Seaweed-associated microbiota have been studied from various scientific perspectives and has revealed dynamic interactions between symbionts and hosts. The bacterial composition of seaweed microbiota has been shown to vary in terms of diversity and abundance, according to host species [74, 75, 76], and even within the same species depending on host tissue type, age, health, and host morphology [76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86]. Furthermore, the environment plays a crucial role in microbial composition and dynamics. For instance, spatial and temporal differences [74, 81, 82, 87, 88], as well as temperature and salinity variations [89, 90, 91], latitude [92], and ocean warming have been shown to influence the microbiota of seaweeds [88]. Finally, the composition of the microbiota differs profoundly from the surrounding water. Microbes form diverse biofilms on the surface of the seaweeds, which constitutes a highly selective habitat [74, 76, 78, 83, 92, 93, 94, 95].
As we acquire more information on the composition of seaweed-associated microbiota, whether it is bacterial, fungal, or other, we also gradually increase our awareness of the intricate interactions that likely exist between algal hosts and their microbiota, as well as between the different members of the microbiota. Seaweeds provide a favorable environment for the proliferation of microorganisms and the formation of biofilms, notably due to the presence of carbon-rich constituents in their cell wall. These constituents serve as sources of nutrients for members of the microbiota [19, 20, 96, 97, 98, 99, 100]. On the other hand, seaweeds may impede microbial growth and biofilm formation through various mechanisms. Several brown seaweeds have, for instance, been shown to react to elicitation with oligoguluronates (oligosaccharides liberated during brown algal cell-wall degradation that are perceived as signal), through oxidative bursts which could serve as an algal defense against bacteria or other colonizers [101, 102]. In this regard, the release of specific organic compounds, such as halogenated or phenolics-related compounds, is noteworthy, given that certain species of kelp (e.g., Nereocystis, Macrocystis, Laminaria, Saccharina) are known to feature a unique iodine-based defense metabolism [103] and phlorotannins present in the Fucales exhibiting antimicrobial properties [104]. To date, only partial information exists on the response of bacteria to particular algal defense compounds, such as in the model bacterium Zobellia galactanivorans, where L-2-haloacid dehalogenases have been shown to increase bacterial tolerance to haloacetic acids [105]. One hypothesis is that the selective pressure and the resulting stability of associations have enabled the evolution of stable, mutually beneficial interactions between hosts and their microbiota. Several prominent model systems demonstrate the extent and capacity of seaweed-dependent microbiota in all major algal lineages. For instance, the green alga Ulva compressa requires at least two growth hormones from its bacterial symbionts—thallusin and a yet unknown compound—to reach its normal growth and morphology [106]. The red alga Delisea pulchra is a model system to study the emergence of dysbiosis in the algal biofilm, but recent work has also shown that some members of the microbiota, such as Phaeobacter sp. BS52, may provide protection against dysbiosis-related changes in the microbiota [107]. Lastly, in the brown alga Ectocarpus subulatus, the bacterial community is essential during host acclimation to low salinity [90]. Metatranscriptomic analyses suggest that the provision of vitamins essential for growth and the regulation of dysbiotic behavior in the bacterial community may be two important features explaining this [108]. Well-studied cases such as these are still an exception in seaweeds, but it is plausible that such interactions are widespread among multicellular eukaryotes and their associated microbiota. Despite criticisms of the term due to the challenge of experimentally demonstrating co-evolution and co-adaptation [109], the significance of host–microbiota interactions in most model systems is now widely acknowledged.
Among seaweeds, Fucaceae are interesting in terms of their microbiota because several species of this family are consistently associated with endophytic fungi (see below for more details). Despite initial groundbreaking research and recent heightened scientific interest in the broader topic of host–microbe interactions, there are still extensive gaps in our knowledge of the Fucaceae microbiota and its interactions with its hosts. In the following section, we will summarize the data currently available in this field and highlight key gaps and challenges toward generating a holistic understanding of these associations.
3.1. The bacterial composition of Fucaceae holobionts
Bacteria, comprising both epi- and endobacterial communities, are an important component of the Fucaceae microbiota. Most information is currently available for the genus Fucus and especially for F. vesiculosus (Table 2). In addition to environmental factors affecting microbial Fucus spp. communities [76, 87, 89, 92, 110, 111], Parrot et al. [83] also described variations in the bacterial communities according to different thallus parts of the host.
Description of bacterial diversity of the Fucaceae from the literature. Only the most abundant taxa are reported
| Algae | Phylum | Class | Family | Genus | Approach | Host localization | Zone | Ref |
|---|---|---|---|---|---|---|---|---|
| Ascophyllum nodosum | Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes | Bacilli, Actinomycetes, Gammaproteobacteria, | Bacillaceaea, Nocardiaceae, Oceanospirillaceae | Bacillus, Rhodococcus, Cobetia, Pseudoalteromonas | Cultivable | Endophyte | Roscoff, France and Oban, Scotland | [112] |
| Proteobacteria, Bacteroidetes, Firmicutes | Alphaproteobacteria, Gammaproteobacteria Betaproteobacteria, Flavobacteriia | Flavobacteriaceae, Oceanospirillaceae, Rhodobacteraceae | Cobetia, Marinomonas, Pseudoalteromonas, Cellulophaga | Cultivable | Epiphyte | Roscoff, France | [75] | |
| Proteobacteria, Bacteroidetes, Actinobacteria | Gammaproteobacteria, Bacteroidota, | Flavobacteriaceae, Alteromonadaceae, Microbacteriaceae, Micrococcaceae | Winogradskyella, Marinobacter Microbacterium, Micrococcus | Cultivable | Epiphyte | Galway Bay, Ireland | [113] | |
| Proteobacteria, Firmicutes, Bacteroidota, Cyanobacteria | Metabarcoding | Epiphyte | Kerry, Ireland | [114] | ||||
| Proteobacteria, Planctomycetes, Actinobacteria, Verrucomicrobia, Cyanobacteria, Bacteroidetes, Firmicutes | Metabarcoding | Epiphyte | Galway Bay, Ireland | [113] | ||||
| Fucus vesiculosus | Proteobacteria, Bacteroidetes, Verrucomicrobia, Cyanobacteria | Gammaproteobacteria, Alphaproteobacteria, | DGGE + Metabarcoding | Epiphyte | Kiel fjord | [87] | ||
| Proteobacteria, Bacteroidetes, Cyanobacteria, | Alphaproteobacteria, Gammaproteobacteria, Deltaproteobacteria | Metabarcoding | Epiphyte | Kiel fjord | [115] | |||
| Proteobacteria, Bacteroidetes | Alphaproteobacteria, Flavobacteriia, Saprospiria | Rhodobacteraceae, Flavobacteriaceae, Saprospiraceaea | Metabarcoding | Epiphyte | Kiel Bight, Western Baltic | [89] | ||
| Proteobacteria | Alphaproteobacteria, Gammaproteobacteria, Flavobacteriia, Saprospiria | Rhodobacteriaceae, Flavobacteriaceae, Saprospiraceaea | Metabarcoding | Epiphyte | Kiel Fjord | [116] | ||
| Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria | Flavobacteria, Bacilli, Actinobacteria, Alphaproteobacteria, Gammaproteobacteria, | Cultivable | Epiphyte | Western Baltic Sea, Germany | [110] | |||
| Proteobacteria, Planctomycetota | Alphaproteobacteria, Planctomycetia, unclassified | Rhodobacteraceae, Erythobacteraceaea, Hyphomonadaceaea | Erythrobacter, Altererythrobacter, Roseobacter | Metabarcoding | Epiphyte | Bülker Leuchtturm | [117] | |
| Fucus vesiculosus | Proteobacteria, Bacteroidetes, Actinobacteria, Cyanobacteria | Alphaproteobacteria, Gammaproteobacteria, Flavobacteriia, Saprospirae, Actinobacteria | Metabarcoding | Epiphyte | Baltic | [88] | ||
| Proteobacteria, Bacteroidetes, Verrucomicrobia | Alphaproteobacteria, Gammaproteobacteria, Bacteroidia | Rhodobacteraceae, Burkholderiaceae, Thiohalorhabdaceae, | Granulosiccocus, Rubritalea, Rhodobacteraceaea_NA, Pleurocapsa, Litorimonas, Blastopirellula | Metabarcoding | Mixed | Western and Eastern Atlantic Ocean | [92] | |
| Proteobacteria, Bacteroidetes | Gammaproteobacteria, Bacteroidia, Alphaproteobacteria | Metabarcoding | Mixed | Acadia National Park, United States | [76] | |||
| Fucus distichus | Bacteroidota, Proteobacteria, Verrucomicrobia, | Flavobacteriales, Alphaproteobacteria, Verrucomicrobiae, Saprospiria, Gammaproteobacteria | Flavobacteriaceae, Rhodobacteraceae, Rubritaleaceae, Saprospiraceae, Thiohalorhabdaceae | Metabarcoding | Epiphyte | Calvert Island, British Columbia, Canada | [111] | |
| Proteobacteria, Bacteroidetes | Gammaproteobacteria, Bacteroidia, Alphaproteobacteria | Metabarcoding | Mixed | Acadia National Park, United States | [76] | |||
| Fucus spiralis | Proteobacteria, Bacteroidetes | Gammaproteobacteria, Alphaproteobacteria, Flavobacteriia, Sphingobacteriia | Rhodobacteraceae, Halomonadaceae, Flavobacteriaceae | Sulfitobacter, Loktanella, Octadecabacter | Metabarcoding | Epiphyte | North Sea, Germany | [118] |
| Proteobacteria, Bacteroidetes | Gammaproteobacteria, Bacteroidia, Alphaproteobacteria | Metabarcoding | Mixed | Acadia National Park, United States | [76] | |||
| Pelvetia canaliculata | Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes | Gammaproteobacteria, Actinomycetes, Bacilli | Pseudoalteromonadaceae, Nocardiaceae, Bacillaceaea, Oceanospirillaceae | Pseudoalteromonas, Rhodococcus, Bacillus, Cobetia | Cultivable | Endophyte | Roscoff, France and Oban, Scotland | [112] |
The bacterial population of A. nodosum was studied long time ago by lab-based culturing [119] and electronic microscopy [120]. Following this, coculture experiments in axenic conditions with the endophytic fungus Mycophycias ascophylli suggested that bacteria protect A. nodosum from being digested by this fungus [121]. After a long gap, the bacterial composition of A. nodosum was further described in 2015, but few studies are available to date. Two of them [113, 114] describe the changes in the epibacterial communities in response to stress, while two others focused on the cultivable bacteria: Martin et al. [75] describe the polysaccharide-degrading activity of surface-attached bacteria, and Tourneroche et al. [112] sought to understand the role of endobacteria in the quorum-sensing system. This latter study also examined the cultivable microbiota of P. canaliculata, constituting the only microbiota study in this genus so far [112]. To our knowledge, as of today, no microbial analyses exist for Pelvetiopsis, Silvetia, and Hesperophycus.
A common point of the above studies was that Proteobacteria and Bacteroidetes were abundant. Furthermore, Firmicutes and Actinobacteria were detected in six studies. One common point among some of these studies is that the presence of several members of the Rhodobacteraceae was positively correlated to abiotic stressors in the environment [78, 88, 89, 116, 118]. However, given the low number of currently available datasets, it is difficult to conclude on general trends, such as the recurring presence of core genera systematically associated with Fucaceae. This would require sampling a broader range of species and conditions and consistent taxonomic assignment based on the same databases at the genus level. As previously reported in green algae [93], it is also likely that there is a core set of bacterial functions consistently associated with the microbiome despite profound taxonomic variability. Yet, this can only be assessed to a limited extent based on 16S rRNA gene metabarcoding data. Exploring these functions would require researchers to include metagenomic approaches to complement metabarcoding approaches and to provide genomic data for the uncultivable components of the microbiota. Thus, even though the bacterial compartment is arguably the best-studied part of the Fucaceae microbiota, much research is still required to obtain a comprehensive picture.
3.2. Fungal composition of Fucaceae holobionts
Microscopic fungi asymptomatically colonize the tissues of healthy seaweed. These algicolous fungi produce bioactive metabolites that are able to kill pathogens or exhibit potent quorum-quenching abilities [112], suggesting that their functions in the host might be based on chemical signaling [122].
The best-known symbiotic associations between Fucaceae and fungi have been described in A. nodosum. This seaweed hosts large symbiotic communities including the fungus Mycophycias ascophylli belonging to Mycosphaerellaceae family (formerly Mycosphaerella ascophylli Cotton) [123, 124, 125, 126, 127]. This latter association was previously designated as a mycophycobiosis by Kohlmeyer and Kohlmeyer [127] and was renamed by Garbary and Deckert as a “symbiotum” [125], a term describing the relationship between a photobiont and a mutualistic endophytic fungus [128]. This fungus is also present in the related Fucaceae P. caniculata (L.) and both brown seaweeds do not occur naturally without this symbiotic fungus [126]. Observations with light microscopy show that fungal hyphae are strongly attached to seaweed cell walls [129]. Their greatest concentration has been observed in receptacles, followed by plant apices, the main axis, and the holdfast [95] and their abundance depends on the maturity of tissue. Reproduction of the mycobiont occurs at the same time as reproduction of the host [125]. Mycophycias ascophylli has been shown to impact host development, fitness and protection against desiccation. It also secretes compounds that may deter herbivores, increase the width of the thallus, and modify host physiology under stressful environments by developing rhizoids for better attachment to substrates [130, 131, 132]. These observations suggest potential interest of some compounds produced by this fungus for plant protection against abiotic stress. At present, there are no ITS (Internal Transcribed Spacer) rDNA sequences available for M. ascophylli in GenBank or UNITE databases [133] but a partial sequence of the Large Subunit 28S rRNA gene is available in the NBCI database [134]. More recently, Vallet et al. [122] isolated a similar fungus from A. nodosum and identified it as a member of the Sordariomycete class related to Moheitopora sp. This taxon was also identified in a recent study exploring the diversity of fungal endophytes in P. caniculata [133].
Beyond this particular example, a number of studies have established fungal cultures from other Fucaceae and revealed a wide diversity of symbionts (Table 3): eight strains were identified in P. canaliculata [122, 126, 133], 46 in F. serratus [135, 136, 137], 32 in F. vesiculosus [117, 136, 138, 139], one in F. spiralis [136]), 18 in A. nodosum [122, 126, 136], and 21 in unidentified species of Fucus [139, 140]. Among these 126 fungal strains, most of them belong to Ascomycota (95%) and only a few to Mucoromycota (2.5%) and Basidiomycota (2.5%). The Ascomycota community, in turn, is equally dominated by Dothideomycetes (35%) and Sordariomycetes (34.2%), followed by Eurotiomycetes (22.5%) and Leothiomycetes (5.8%). The use of CARD-FISH to study the tissue of F. vesiculosus also revealed a positive signal for fungi belonging to the class Eurotiomycetes [116]. The majority of these studies have concentrated on characterizing the endophytic algicolous fungi associated with different species of Fucus spp.
Description of fungal diversity of Fucaceae from the literature
| Seaweed | Taxonomy (p_phylum;c_class;f_family;g_genus;s_species) | Approach | Host localization | Zone | Ref |
|---|---|---|---|---|---|
| Pelvetia canaliculata | p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium;s_cladosporioides p_Ascomycota;c_Dothideomycetes;f_Didymellaceae;g_Didymella;s_exigua p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Alternaria;s_metachromatica p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Stemphylium;s_vesicarium p_Ascomycota;c_Sordariomycetes;f_Hypocreomycetidae;g_Acremonium;s_exuvarium p_Ascomycota;c_Sordariomycetes;f_;g_Chlamydocillium;s_cyanophilum | Cultivable | Endo | Wenbry, Devon, UK | [141] |
| p_Ascomycota;c_Dothideomycetes;f_Mycosphaerellaceae;g_Mycosphaerella;s_ascophylli | Cultivable | Endo | College Rocks, Aberystwyth, UK | [126] | |
| p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Paradendryphiella;s_arenariae | Cultivable | Endo | Oban, Scotland, UK | [122] | |
| Ascophyllum nodosum | p_Ascomycota;c_Agaricomycetes;f_Polyporaceae;g_Trametes;s_versicolor p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium;s_cucumerinum p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Paradendryphiella;s_arenariae p_Ascomycota;c_Dothideomycetes;f_Phaeosphaeriaceae;g_Phaeosphaeria p_Ascomycota;c_Dothideomycetes;f_Thyridariaceae;g_Roussoella p_Ascomycota;c_Dothideomycetes;f_;g_Exosporium;s_stylobatum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_brevicompactum p_Ascomycota;c_Leotiomyctes;f_Sclerotiniaceae;g_Botryotinia;s_fuckeliana p_Ascomycota;c_Sordariomycetes;f_Plectosphaerellaceae;g_Verticillium ;s_cf biguttatum p_Ascomycota;c_Sordariomycetes;f_Xylariaceae;g_Brunneiperidium;s_gracilentum p_Ascomycota;c_Sordariomycetes;f_;g_Moheitospora p_Ascomycota;c_Dothideomycetes;f_Microsphaeropsidaceae;g_Microsphaeropsis;s_olivacea | Cultivable | Endo | Roscoff, France & Oban, Scotland, UK | [122] |
| Fucus spiralis | p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium p_Ascomycota;c_Dothideomycetes;f_Dictyosporiaceae;g_Dendryphiella;s_salina p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Aspergillus ;s_fumigatus p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Aspergillus p_Mucoromycota;c_Mucoromycetes;f_Lichtheimiaceae;g_Lichtheimia;s_corymbifera | Cultivable | Endo | West Burra, Shetland Islands, UK & Etete, New Brunswick, Canada | [136] |
| Fucus serratus | p_Ascomycota;c_Dothideomycetes;f_Mycosphaerellaceae;g_Mycosphaerella;s_ascophylli | Cultivable | Endo | College Rocks, Aberystwyth, UK | [126] |
| p_Ascomycota;c_Ascomycetes;f_Myxotrichaceae;g_Oidiodendron p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Acroconidiella p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium p_Ascomycota;c_Dothideomycetes;f_Coniothyriaceae;g_Coniothyrium p_Ascomycota;c_Dothideomycetes;f_Dictyosporiaceae;g_Dendryphiella;s_salina p_Ascomycota;c_Dothideomycetes;f_Didymellaceae;g_Epicoccum p_Ascomycota;c_Dothideomycetes;f_Mycospaerellaceae;g_Mycosphaerella p_Ascomycota;c_Dothideomycetes;f_Periconiaceae;g_Periconia p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Alternaria p_Ascomycota;c_Dothideomycetes;f_;g_Phoma p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Aspergillus p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium p_Ascomycota;c_Eurotiomycetes;f_Herpotrchiellaceae;g_Phialophora p_Ascomycota;c_Eurotiomycetes;f_Thermoascaceae;g_Paecilomyces p_Ascomycota;c_Leotiomyctes;f_Sclerotiniaceae;g_Botrytis;s_cinerea p_Ascomycota;c_Sordariomycetes;f_Apiosporaceae;g_Arthrinium p_Ascomycota;c_Sordariomycetes;f_Chaetomiaceae;g_Chaetomium p_Ascomycota;c_Sordariomycetes;f_Chaetomiaceae;g_Humicola;s_fuscoatra p_Ascomycota;c_Sordariomycetes;f_Halosphaeriaceae;g_Corollospora;s_angusta p_Ascomycota;c_Sordariomycetes;f_Hypocreaceae;g_Trichoderma p_Ascomycota;c_Sordariomycetes;f_;g_Acremonium p_Ascomycota;c_Sordariomycetes;f_;g_Emericellopsis;s_minima p_Ascomycota;c_Sordariomycetes;f_Nectriaceae;g_Fusarium p_Ascomycota;c_Sordariomycetes;f_Microascaceae;g_Scopulariopsis p_Ascomycota;c_Sordariomycetes;f_Plectosphaerellaceae;g_Verticillium;s_cinnabarinum p_Ascomycota;c_Sordariomycetes;f_Xylariaceae;g_Nodulisporium p_Mucoromycota;c_Mucoromycetes;f_Mucoraceae;g_Mucor p_Ascomycota;c_Dothideomycetes;f_Didymellaceae;g_Epicoccum;s_purpurascens p_Ascomycota;c_Leotiomyctes;f_Dematiaceae;g_Asteromyces;s_cruciatus p_Ascomycota;c_Leotiomyctes;f_Pseudeurotiaceae;g_Geomyces p_Ascomycota;c_Leotiomyctes;f_;g_Tetracladium;s_maxilliformis p_Ascomycota;c_Sordariomycetes;f_Chaetomiaceae;g_Chaetomium;s_funicola p_Ascomycota;c_Sordariomycetes;f_Diaporthaceae;g_Phomopsis p_Ascomycota;c_Sordariomycetes;f_Halosphaeriaceae;g_Corollospora;s_intermedia p_Ascomycota;c_Sordariomycetes;f_Halosphaeriaceae;g_Sigmoidea;s_marina p_Ascomycota;c_Sordariomycetes;f_Hypocreaceae;g_Gliocladium p_Ascomycota;c_Sordariomycetes;f_;g_Acremonium;s_mumorum p_Ascomycota;c_Sordariomycetes;f_;g_Acremonium;s_tubakii p_Ascomycota;c_Sordariomycetes;f_;g_Acremonium;s_fuci p_Ascomycota;c_Sordariomycetes;f_Lulworthiaceae;g_Lindra;s_obtusa p_Ascomycota;c_Sordariomycetes;f_Microascaceae;g_Microascus | Cultivable | Mixed | Heligoland Island, Germany | [137] | |
| p_Ascomycota;c_Dothideomycetes;f_Coniothyriaceae;g_Coniothyrium p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Aspergillus ;s_fumigatus p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium | Cultivable | Endo | West Burra, Shetland Islands, UK | [136] | |
| Fucus vesiculosus | p_Ascomycota;c_Dothideomycetes;f_;g_Phoma p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_chrysogenum | Cultivable | Epi | Kiel Fjord, Baltic Sea, Germany | [117] |
| p_Ascomycota;c_Dothideomycetes;f_Pyrenochaetopsidaceae;g_Pyrenochaetopsis p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_atrovenetum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_biourgeianum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_canescens p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_glabrum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_sanguifluum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_spathulatum p_Ascomycota;c_Leotiomyctes;f_Ploettnerulaceae;g_Cadophora;s_malorum p_Ascomycota;c_Sordariomycetes;f_;g_Acremonium;s_furcatum p_Ascomycota;c_Sordariomycetes;f_;g_Stilbella;s_fimeteria p_Ascomycota;c_Sordariomycetes;f_Hypocreaceae;g_Trichoderma;s_gamsii p_Ascomycota;c_Sordariomycetes;f_Hypocreaceae;g_Trichoderma;s_paraviridescens p_Ascomycota;c_Sordariomycetes;f_Nectriaceae;g_Fusarium;s_graminearum p_Ascomycota;c_Sordariomycetes;f_Plectosphaerellaceae;g_Gibellulopsis p_Ascomycota;c_Sordariomycetes;f_Plectosphaerellaceae;g_Gibellulopsis;s_nigrescens p_Ascomycota;c_Sordariomycetes;f_;g_Emericellopsis p_Ascomycota;c_Sordariomycetes;f_;g_Emericellopsis;s_terricola p_Ascomycota;c_Wallemiomycetes;f_Wallemiaceae;g_Wallemia;s_muriae | Cultivable | Endo | Kiel Fjord, Baltic Sea, Germany | [117] | |
| p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_brevicompactum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_coralligerum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium | Cultivable | Mixed | Kiel Fjord, Baltic Sea, Germany | [117] | |
| p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Aspergillus ;s_fumigatus p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium p_Ascomycota;c_Leotiomyctes;f_Pezizellaceae;g_Chalara p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Alternaria p_Ascomycota;c_Dothideomycetes;f_Coniothyriaceae;g_Coniothyrium | Cultivable | Endo | West Burra, Shetland Islands, UK | [136] | |
| p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Dendryphiella;s_salina p_Ascomycota;c_Dothideomycetes;f_;g_Phoma p_Ascomycota;c_Sordariomycetes;f_Ophiocordycipitaceae;g_Tolypcladium;s_inflatum | Cultivable | Endo | Lulworth Coast, UK & Cuxhaven, Germany | [139] | |
| p_Ascomycota;c_Dothideomycetes;f_Didymellaceae;g_Epicoccum | Cultivable | Endo | Tönning, Germany | [138] | |
| Fucus sp. | p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium;s_europaeum p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium;s_ramotenellum p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium;s_sphaerospermum p_Ascomycota;c_Dothideomycetes;f_Cladosporiaceae;g_Cladosporium p_Ascomycota;c_Dothideomycetes;f_Didymellaceae;g_Epicoccum;s_nigrum p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Alternaria p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Aspergillus p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium;s_brevicompactum p_Ascomycota;c_Eurotiomycetes;f_Aspergillaceae;g_Penicillium p_Ascomycota;c_Eurotiomycetes;f_Herpotrichiellaceae;g_Exophiala p_Ascomycota;c_Sordariomycetes;f_Bionectriaceae;g_Clonostachys;s_rosea p_Ascomycota;c_Sordariomycetes;f_Cordycipitaceae;g_Engyodontium p_Ascomycota;c_Sordariomycetes;f_Hypocreaceae;g_Trichoderma;s_paraviridescens p_Basidiomycota;c_Cystobasidiomycetes;f_Symmetrosporaceae;g_Symmetrospora p_Basidiomycota;c_Microbotryomycetes;f_;g_Leucosporidium p_Basidiomycota;c_Tremellomycetes;f_Cryptococcaceae;g_Cryptococcus p_Mucoromycota;c_Mucoromycetes;f_Mucoraceae;g_Rhizopus;s_orysae | Cultivable | Mixed | Scheveningen, Netherlands | [140] |
| p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Dendryphiella;s_arenariae p_Ascomycota;c_Dothideomycetes;f_Pleosporaceae;g_Dendryphiella;s_salina p_Ascomycota;c_Dothideomycetes;f_;g_Phoma p_Ascomycota;c_Sordariomycetes;f_Apiosporaceae;g_Arthrinium | Cultivable | Endo | Lulworth Coast, UK & Tönning, Travemünde, Cuxhaven, Germany | [139] |
To date, only two studies have explored the non-cultivable fungal diversity of Fucaceae by DNA fingerprinting methods [133, 137]. The fungal endophytes of P. canaliculata are mainly composed of Ascomycota, including one abundant unclassified Amplicon Sequence Variant (ASV), and of Basiodiomycota. In other studies of the P. canaliculata- or A. nodosum-associated microbiota, the endophyte M. ascophylli remained unidentified either in culture or in fungal NGS databases [133]. The identity of fungi colonizing F. serratus has been explored through Denaturing Gradient Gel Electrophoresis (DGGE) analysis. These studies indicate that understanding the ecological role and deciphering the core fungal composition in seaweeds is challenged by a lack of knowledge of the non-cultivable fungal diversity. Indeed, there is a limited amount of available reference ITS rDNA sequences of fungal taxa from marine environments. There is thus an urgent need for the development of culture collections, the description of new species, and the sequencing of specimens, particularly those that are not amenable to culture. The development of new tools such as Mass Imaging Spectrometry (MIS) associated with CARD-FISH are also useful to further investigate the fungal diversity associated with its metabolic potential and to decipher the molecular interactions involved in seaweed–fungal symbiosis.
3.3. Archaeal and viral community associated with Fucaceae: the great unknown
Unlike fungi and bacteria, to date no study on the archaeal or viral partners of the Fucaceae holobiont is available in the literature, but this does not indicate their absence or a lack of importance. While archaea were initially discovered in extreme environments, we now know that they are widely distributed in marine environments [142]. Among the four phylogenetic groups (i.e., the Euryarchaeota phylum and the TACK, Asgardarchaeota, and DPANN superphyla), archaea of the DPANN superphylum are commonly found living in symbiosis, frequently with other archaea or bacteria [143]. However, most metabarcoding studies examining alga-associated microbiota (see above) use bacteria-biased primers and remove any sequences potentially resembling archaea during the usual assembly pipeline although suitable generic primers now exist [144]. Even when looking beyond the Fucaceae to other brown algae, archaeal diversity on brown algae has so far only been examined in one study. Trias et al. [145] used primers targeting the amoA gene combined with a cloning approach to assess the diversity of ammonium-oxidizing bacteria and archaea in Laminaria rodriguezii, and revealed the presence of a diverse archaeal community.
Similar observations can also be made for viruses. It is generally accepted that viruses infect virtually all living organisms including brown algae [146]. Much of the work on brown algal viruses has focused on Phaeoviruses, large double-stranded DNA viruses that infect wall-less gametes or spores [147]. The most prominent representative of this group is arguably Ectocarpus siliculosus virus 1 (EsV-1), which was also found integrated into its host’s genome [148] but representatives of this group have also been found in other Ectocarpales [149] and in kelp [150, 151]. Other studied brown algal viromes are known from the kelp Ecklonia radiata and that of the Scytosiphon lomentaria (Ectocarpales). In the former, numerous members of Siphoviridae, Myoviridae and other viruses were detected, including a CRESS DNA virus-related sequence found specifically in bleached individuals [152]. In the latter, two Endornaviridae and two unclassified RNA viruses were sequenced, but no information about their interactions with the host is available [153]. However, within the Fucaceae no virome studies have been published so far, although one study reported that F. serratus expresses a range of RNA Interference-like responses to viruses [154], suggesting that there is regular contact with viruses in Nature. Indeed, viromes may be highly dynamic, and shifts may occur in response to stress, as shown in the brown alga Ectocarpus [154]. Furthermore, host-associated viromes also include phages, which interact directly with the bacterial components of the alga-associated microbiota and as such affect interactions [155].
This lack of knowledge on archaea and viruses may considerably limit our capacity to understand the full spectrum of processes occurring within Fucaceae holobionts. We thus undeniably require more data on both to improve our understanding of the interactions within the Fucaceae holobionts.
4. Microbiota-derived compounds and their biological activities
Host-associated microbiota, whether they are endophytes or part of the biofilm (i.e., epiphytes), are likely to interact with their hosts in various ways and frequently these interactions are based on their capacity to produce specific essential metabolites for algal holobiont [112, 122, 156, 157]. In the following section, we will provide an overview of the current knowledge of the chemical landscape of algal partners, highlighting key specialized metabolites with potential applications in different economic sectors.
4.1. Bacterial metabolites
Most of the reported secondary metabolites from bacteria associated with Fucaceae have been derived from strains isolated from the surface of Fucus and notably the genus Roseobacter. This particular genus of bacteria is renowned for its capability to synthetize N-Acyl Homoserine Lactones (AHLs), which are well-known bacterial Type 1 Autoinducer signal compounds used by Gram-negative bacteria for Quorum Sensing (QS). QS is a cell density-dependent mechanism regulating physiological traits such as antibiotic production, virulence, motility, bioluminescence, or biofilm formation [158, 159]. The AHLs isolated from Roseobacter are commonly made up of a 𝛾-lactone ring and an acyl side chain which is usually even-numbered and unbranched, ranging in chain length from C4 to C18. A sampling campaign of Roseobacter from F. spiralis harvested in the German Wadden Sea and in the eastern North Sea led to the isolation of several marine strains, including Roseovarius sp., Loktanella sp., Octadecabacter sp., and Dinoroseobacter sp. In 2019, Ziesche et al. [160] isolated known AHLs (Figure 3.1, 2, 5–18) and identified two original ones (3, 4), and their structures were confirmed by synthesis [160, 161, 162, 163, 164]. All of these compounds displayed a high QS activity in biosensor models. High variability in the composition of AHLs has been observed among strains from different sample locations. For instance, surface-associated strains produce AHLs more frequently compared to strains isolated from sediments or from the water column [160].
Chemical structures of seaweed-associated bacterial compounds (1–45). All these compounds have been classified according to their putative metabolic pathway: Polyketide Synthase (PKS), Fatty acids and structurally modify fatty acids, Alkaloids, Shikimates and Phenylpropanoids, and miscellaneous.
Chemical studies of the strains Loktanella sp. and Roseovarius sp. isolated from F. spiralis led to the isolation of seventeen related N-acylalanine methyl esters (NAMEs) (19–34, 36) [165]. Seven N-acylglycine methyl esters (NAGMEs) (19–25) were extracted from Loktanella sp., nine N-acylalanine methyl esters (NAMEs) (26–34) and one N-acyl-2-aminobutyric acid methyl ester (NABME) (36) from Roseovarius sp. Compounds 35 (NABME derivative) and 37 (N-acylvaline methyl ester derivative—NAVME) were also isolated from bacterial strains found in an unidentified species of Fucus [165]. Despite their structural analogy with AHLs, no QS activity was found, thus suggesting that they play a different ecological role. Compound 36 displayed some antibacterial activities and compound 35 had both antibacterial and cytotoxic properties (Supplementary Table S1).
Phaeochromycin E, C, and B (respectively 38, 39, 40) along with the chroman derivative (41) were isolated from the endophytic strain Streptomyces sundarbansensis associated with a species of Fucus collected from the coastline of Algeria [166]. These compounds showed anti-inflammatory and antibacterial activities [166, 167]. Furthermore, the stilbene Kocumarin (42) isolated from the actinobacterium Kocuria marina associated with P. canaliculata [168] displayed antibacterial effects [168, 169].
In 2019, the first study based on the surface microbiota of F. vesiculosus combining Mass Spectrometry Imaging (MSI), metabolomics, and dereplication allowed for the putative annotation of compounds produced by Streptomyces spp. such as Obelmycin F (43), Azinomycin B (44), and Indolmycin (45) and their localization on the thallus and tips of the algal tissues [83]. Compounds 43 and 45 show both antibacterial activities and bacteriostatic effects, while compound 44 is known for its antitumor activity [170, 171, 172]. This MSI approach also demonstrated the presence of AHLs on the surface of the seaweed, although it cannot identify the organisms responsible for its production.
Altogether the capacity of these Fucaceae-associated strains to produce antibacterial and cytotoxic compounds likely impacts both the host and other members of the microbiota, but also harbors great potential for numerous industrial applications as described in Section 2.
4.2. Fungal metabolites
Fungi have proven to be a highly diverse yet understudied reservoir of biologically active compounds exhibiting a plethora of potential functions and applications. Here we present a non-exhaustive list of secondary metabolites isolated from fungal strains associated with the Fucaceae.
Chemical investigation of the fungus Paradendryphiella salina, isolated from the marine brown alga A. nodosum and P. canaliculata, led to the identification of the dendryphiellones A and D (Figure 4.46–47) [173], which have the ability to inhibit the AHL-mediated QS. In the same way, extracts of fungal endophytes have been found to inhibit the auto inducer 2 (AI-2) pathway, another QS-signaling pathway known to be involved in cross-kingdom signaling [112]. As QS is thought to be responsible for a significant increase in the expression of virulence-related genes in aquaculture farms [174], compounds 46 and 47 have been suggested as possible agents to target specific bacterial pathogens [173] and could be interesting as aquaculture feed additives.
Chemical structures of seaweed-associated fungal compounds (46–82). All compounds have been classified according to their putative metabolic pathway: Polyketide Synthase (PKS), Non-Ribosomal Peptide synthetase (NRPS), Terpenoids.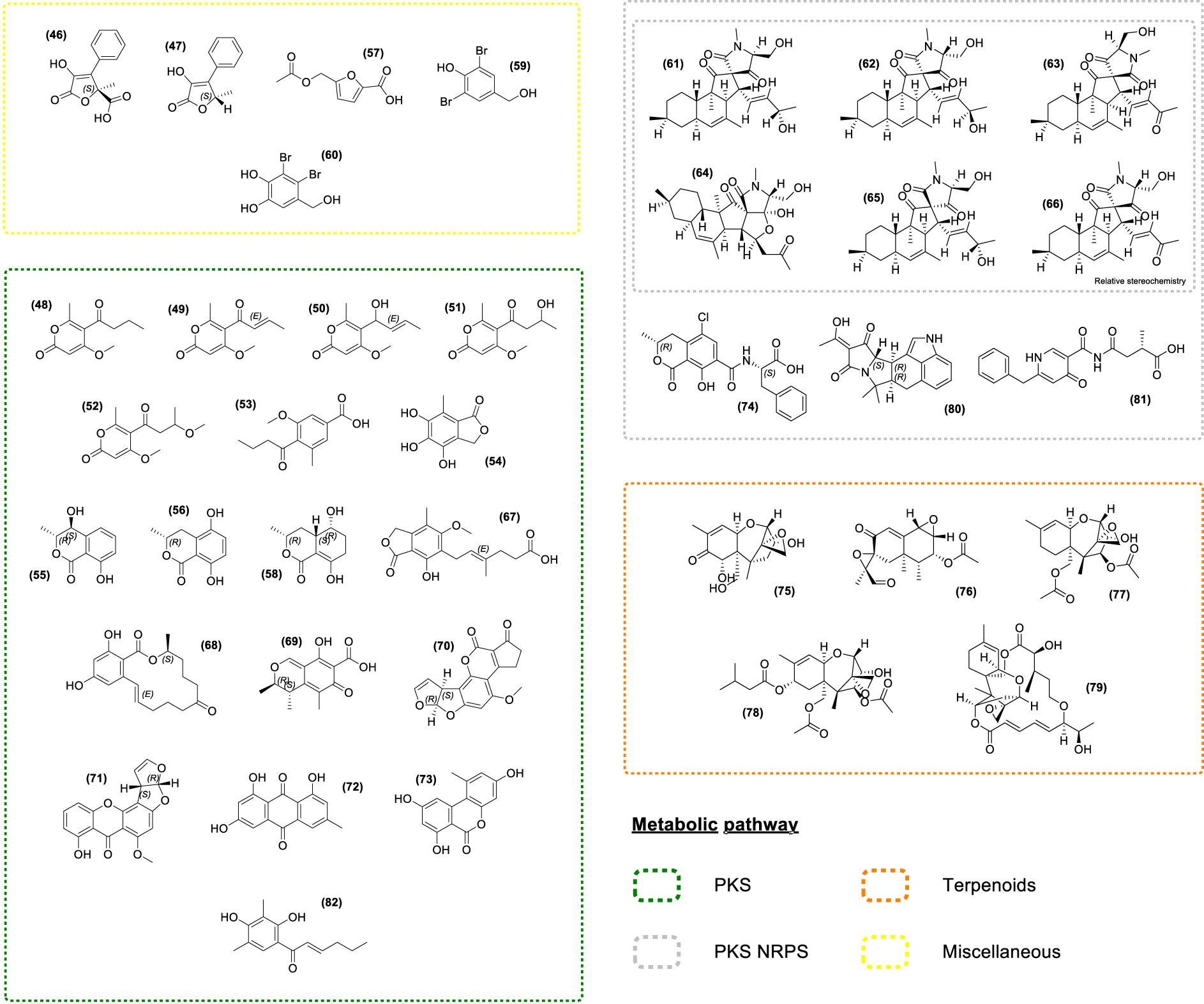
The endophytic fungus Phaeosphaeria sp. isolated from A. nodosum led to the discovery of several polyketides including the new Pyrenocine S (48) along with known Pyrenocines (49, 50, 51, 52) and Pyrenochaetic acid C (53) [122]. Most of the Pyrenocines described to date are effective in inhibiting infection by protistan pathogens in the brown alga Ectocarpus siliculosus. Strikingly, these compounds also inhibited the infection of nori (Pyropia yezoensis) by its two most devastating oomycete pathogens, Olpidiopsis pyropiae and Pythium porphyrae [122]. Several polyketides were also identified in other fungal strains. For instance, the fungus Epicoccum sp. isolated from F. vesiculosus produces Epicoccone A (54), (3R,4S)-4-hydroxymellein (55), (3R)-5-hydroxymellein (56), and acetyl Sumiki’s acid (57) [138]. The ensuing biological evaluation pointed out the antibacterial activity of the acetyl Sumiki’s acid (57) [175] while Epicoccone A (54) displayed antioxidant, antibacterial, and cytotoxic properties [176, 177, 178], making these metabolites potential candidates for cosmetics and pharmaceutical industries (Supplementary Table S1).
The derivatives of the ubiquitous mellein, (55) [155, 156, 157, 158, 159, 160] and (56) have numerous interesting properties such as cytotoxic, antibacterial, antitrypanosomal activities as well as insecticidal properties [179, 180, 181, 182, 183, 184, 185, 186, 187]. 5-hydroxyramulosin (58), a polyketide analog of 55 and 56 was isolated from the endophytic fungus Phoma tropica associated with F. spiralis and showed activities in a range of biological assays [188]. It displayed a potent cytotoxic activity along with some antifungal and antibacterial effects [189, 190].
3,5-dibromo-p-hydroxybenzyl alcohol (59) and Lanosol (60) were isolated from a non-assigned marine Ascomycetes sp. from A. nodosum [132]. Lanosol (60) was also detected in the brown algae F. vesiculosus and A. nodosum [132, 191]. Thus, the possibility that marine Ascomycetes living as epi- or endophytes in these brown algae were producing these bromophenols cannot be neglected. Both compounds displayed antibacterial effects [191] and their halogenated nature suggests antifouling properties. Indeed, red algae cultivated in a bromide-free environment exhibited increased bacterial colonization in comparison with cultivation in a bromide-containing medium [192].
Chemical exploration using a bioactivity-driven molecular networking approach on the endophytic fungi Pyrenochaetopsis sp. from F. vesiculosus led to the identification of six new hybrid polyketides, thought to be synthesized by polyketide synthases/non-ribosomal peptide synthetases (PKS–NRPS) [193, 194, 195], Pyrenosetins A-F (61–66). They have antineoplastic and cytotoxic activities, except for Pyrenosetin E (65) which shows only cytotoxic activity, and Pyrenosetin F (66) bearing no biological activity on the assayed targets [193, 194, 195].
The study of the toxic metabolites produced by species of Fusarium, Alternaria, Penicillium, and Aspergillus associated with A. nodosum, P. canaliculata, and F. vesiculosus demonstrated the presence of numerous mycotoxins on the surface of the thallus of these three seaweeds [196]. Among them, polyketides such as Mycophenolic acid (67), Zearalenone (68), Citrinin (69), Aflatoxin B1 (70), Sterigmatocystin (71), and Emodin (72) along with Alternariol (73) and Ochratoxin A (74) were found. Moreover, other common mycotoxins were detected, such as the Deoxynivalenol (75), PR toxin (76), Diacetoxyscirpenol (77), T-2 toxin (78), Roridin A (79) and 𝛼-Cyclopiazonic acid (80). Indeed, species of the family Fucaceae show a broad profile of mycotoxins, while no mycotoxins were found on L. digitata and S. latissima surfaces from the family Laminariaceae [196]. Lastly, Pestalamide B (81) and Dihydrosorbicillin (82) were detected by MSI in F. vesiculosus tissues [83]. Of these two compounds, only Pestalamide B has antifungal activity against Aspergillus fumigatus [197].
Together, these results suggest that fungal endophytes associated with Fucaceae produce a wide range of bioactive metabolites which might confer protection against pathogen infection or prevent dysbiosis in the phycosphere, i.e., microscale area around phytoplankton cells enriched in organic substrates [198]. However, few studies have reported on the potential ecological roles of isolated metabolites. Indeed, most of the studies looked for potential biological activities of the fungal compounds and are thus largely confined to classical in vitro laboratory assays. The in situ study of chemical interactions within algae is a key element to help understand the chemical ecology of the holobiont as a whole. Inoculations of seaweeds with known microbiota are necessary to transpose studies on isolated microorganisms to the holobiont community. Such multivariable and integrative experiments are still in their infancy but new tools such as MiSCoTo [199] allow predicting the metabolic networks of a host and members of the microbiota, and these models have some predictive power for metabolite profiles of the “reconstituted” holobiont observed in vitro [200].
5. Conclusions and perspectives
We have shown here the huge diversity of the bacterial and fungal communities associated with members of the Fucaceae, as well as the chemical diversity of metabolites produced by these microorganisms. Although our knowledge of the microbiota of Fucaceae is still rudimentary, in particular regarding Archaea and viruses, we are starting to notice that some fungal and bacterial species appear repeatedly and in high abundance in the studies conducted so far. One of the challenges in the near future will be to properly define a core microbiota for each algal host, to monitor the factors (i.e., geographical areas and seasons) that may lead to some variations in the holobiont composition, to link these variations to the host fitness, and lastly to decipher the chemical traits which might impact the bioactivities and thus the applications.
As the field of marine chemical ecology continues to grow, we are gaining a more comprehensive understanding of the numerous ecological functions of marine natural products. Recent research has made significant contributions to our understanding of the diverse roles of host-associated bacteria and fungi in marine algal communities. Furthermore, advances in molecular methods have shown that some microorganisms are responsible for the production of specialized metabolites that chemically defend and/or structure the microbiota of the host organism. Chemical communication and sensory ecology are among the main topics in marine ecology that have grown significantly in recent years. Only by bringing together biology, chemistry, bioinformatics, systems biology, as well as bioactivity screening can we start exploring algal holobiont as a whole. Such interdisciplinary approaches and new technologies will play a key role in furthering our understanding of marine chemical ecology. Furthermore, deciphering the chemical role of each partner within the algal holobiont will lead to a better understanding of the origins and functions of the metabolites produced by the holobiont, including some of the compounds of economic interest cited before as well as novel bioactive compounds, the exploration of which is still in its infancy. In the long run, we anticipate this knowledge will then feed back into the harvesting (e.g., choice of ideal harvesting period), production (e.g., use of probiotics to manipulate holobiont metabolomes or direct use of microbes to enrich in the target metabolites) and valorization of the Fucaceae.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.
Funding
This work was funded in part by the SEABIOZ ANR project (AAPG2020, ANR-20-CE43-0013), the ANRT and the Centre Mondial de l’Innovation of the Groupe Roullier (Cifre grant number 2022/0938 to GD), the UMR 8227 of the Roscoff Biological Station (CNRS, Sorbonne University), the UMR 7245 of the National Museum of Natural History.
Acknowledgments
We would like to greatly thank Christelle Parchemin (Centre de Recherches Insulaires et Observatoire de l’Environnement—CRIOBE, Perpignan France) for providing help and support for this work.




 CC-BY 4.0
CC-BY 4.0