1. Introduction
Peptides and proteins can form specific folded structures necessary for their biological functions. This folding is ensured by intermolecular or intramolecular hydrogen bonds, between the carbonyl groups and the amine protons of the amide bond, as well as by electrostatic and hydrophobic interactions of the side chains. Artificial foldamers are repetitive synthetic structures that adopt folded molecular configurations, thus mimicking the structural properties of biomolecules [1]. With the increasing use of therapeutic peptides [2, 3, 4], interest in peptide-based foldamers capable of mimicking secondary structures is growing rapidly [5, 6, 7, 8]. Indeed, peptidomimetic foldamers offer solutions to overcome two main issues of short peptides: their lack of stability against proteolysis and their low ability to adopt specific conformations, thereby limiting their practical use in certain therapeutic fields such as the inhibition of protein–protein interactions.
Fluorinated molecules have generated considerable interest in medicinal chemistry, with approximately 150 reaching the pharmaceutical market and accounting for nearly 30% of all newly approved drugs [9, 10, 11]. Furthermore, incorporating fluorinated amino acids into peptides and proteins has increased their biological activity and propensity to form secondary structures due to fluorine’s ability to enhance local hydrophobicity and significantly strengthen hydrogen bonds [12, 13, 14]. The addition of fluorinated amino acids can also improve chemical and enzymatic stability [11, 12] although in some cases, the effect is less pronounced with direct fluorinated analogues of natural amino acids [14]. Additionally, fluorine labeling is a significant advantage for studying biological events using 19F NMR spectroscopy, such as the structure and function of biomolecules, enzymatic mechanisms, ligand–biomolecule interactions, metabolic pathways, and for 19F MRI medical imaging [15]. However, to our knowledge, very few examples of fluorinated foldamers have been reported [16, 17, 18, 19]. Regarding peptidic foldamers, fluorinated peptoids [18], CF3-Aib [20], CF3-β-peptides [21], and CF3-pseudoprolines [22] adopt helical conformations, while oligomers of cis-2-amino-1-fluorocyclobutane-1-carboxylic acid [16] and 2-fluoro-aryl substituted α,β 2,3-amino acid [17] adopt a strand-like secondary structure. As part of our interest in fluorinated peptidomimetics and foldamer construction, we are currently exploring the potential of fluorinated foldamers. This new class of foldamers, noted by a few chemists to date for their conformational properties [16, 18, 19, 20, 21], remains entirely unknown in medicinal chemistry and as potential drug candidates. Our laboratory has been interested in designing new fluorinated foldamers based on N-difluoromethyl triazole units. As a proof of concept of their benefit in medicinal chemistry, the design of potential modulators of protein aggregation in the field of type 2 diabetes (T2D) was investigated.
T2D is a significant public health challenge, currently affecting around 400 million individuals worldwide. This figure is projected to surpass half a billion by 2045, yet no causal treatment exists for this condition [23, 24]. T2D belongs to the large family of diseases called amyloidosis caused by the pathological aggregation of amyloid proteins [25, 26, 27]. The aggregation of human islet amyloid polypeptide (hIAPP, also known as amylin) plays a role in the death of pancreatic β-cells in T2D patients through a mechanism that is only partially understood. The hIAPP undergoes a conformational change that leads to an aggregation pathway, forming toxic soluble oligomers and fibers. These aggregated species adopt β-sheet rich conformations, and amyloid deposits are found in the pancreas of over 95% of diabetic patients [28, 29, 30]. Preventing both the presence of soluble transient oligomers and the later aggregated forms of hIAPP is essential to mitigate cell toxicity caused by membrane disruption and other less understood phenomena induced by hIAPP aggregation [31, 32, 33]. This approach must be investigated to develop a viable therapy for T2D.
2. Synthesis of aminomethyl-1,4-triazolyl-difluoroacetic acid
To start, we designed and prepared an original fluorinated peptidomimetic by incorporating N-difluoromethyltriazole into its structure. The aminomethyl-1,4-triazolyl-difluoroacetic acid named N-difluoromethyl 1,4-Tz amino acid (1,4-Tz-CF2), N-difluoromethyl 1,4-Tz, or 1,4-Tz-CF2 amino acid can be considered an analogue of Gly-difluoroGly dipeptide, which is unstable [34].
The pseudotetrapeptide was obtained by copper-catalyzed dipolar cycloaddition from an azide and an alkyne carrying amino acid residues. This reaction was carried out in the presence of copper sulfate pentahydrate, sodium ascorbate, and finally TBTA as a ligand (Scheme 1).
Synthesis of N-CF2 triazole.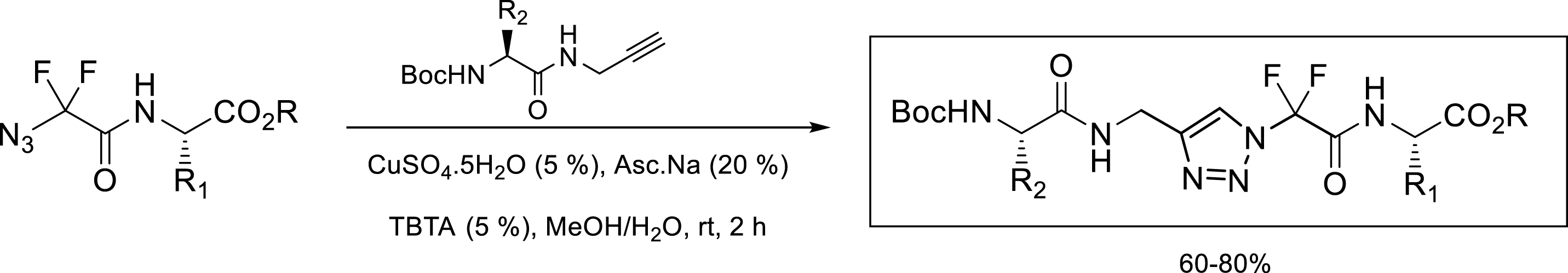
We demonstrated that the introduction of one monomer 1,4-Tz-CF2 in short peptides could induce extended structures. NMR, DFT, and X-ray crystallography showed the interactions between fluorine atoms and neighboring protons [34]. The 2D 19F-1H NOE NMR experiments allowed identifying correlations between the fluorine atoms and the proton of the triazole and the NH of phenylalanine. These observations were in agreement with the X-ray diffraction analyses. The structure identified within the crystal lattice highlights the distances between the two protons (triazole proton/NH phenylalanine) and a fluorine atom, which are 2.58 Å and 2.29 Å with φ angles of 88° and 107°, respectively. This information tends to confirm the proton–fluorine interactions stabilizing an extended local conformation (Figure 1).
Interactions between fluorine atoms and neighboring protons [34].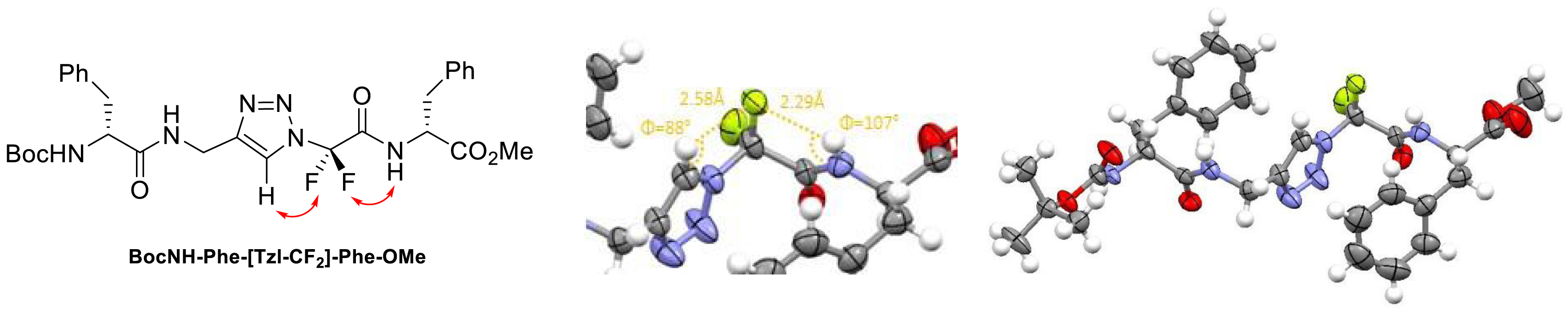
In view of these particular characteristics of this N-CF2-triazole unit, we were interested in incorporating it into oligomers to investigate their specific conformations and their potential as modulators of hIAPP amyloid aggregation. We therefore focused our attention on different and original classes of fluorinated triazolamers.
3. Synthesis of homotriazolamers
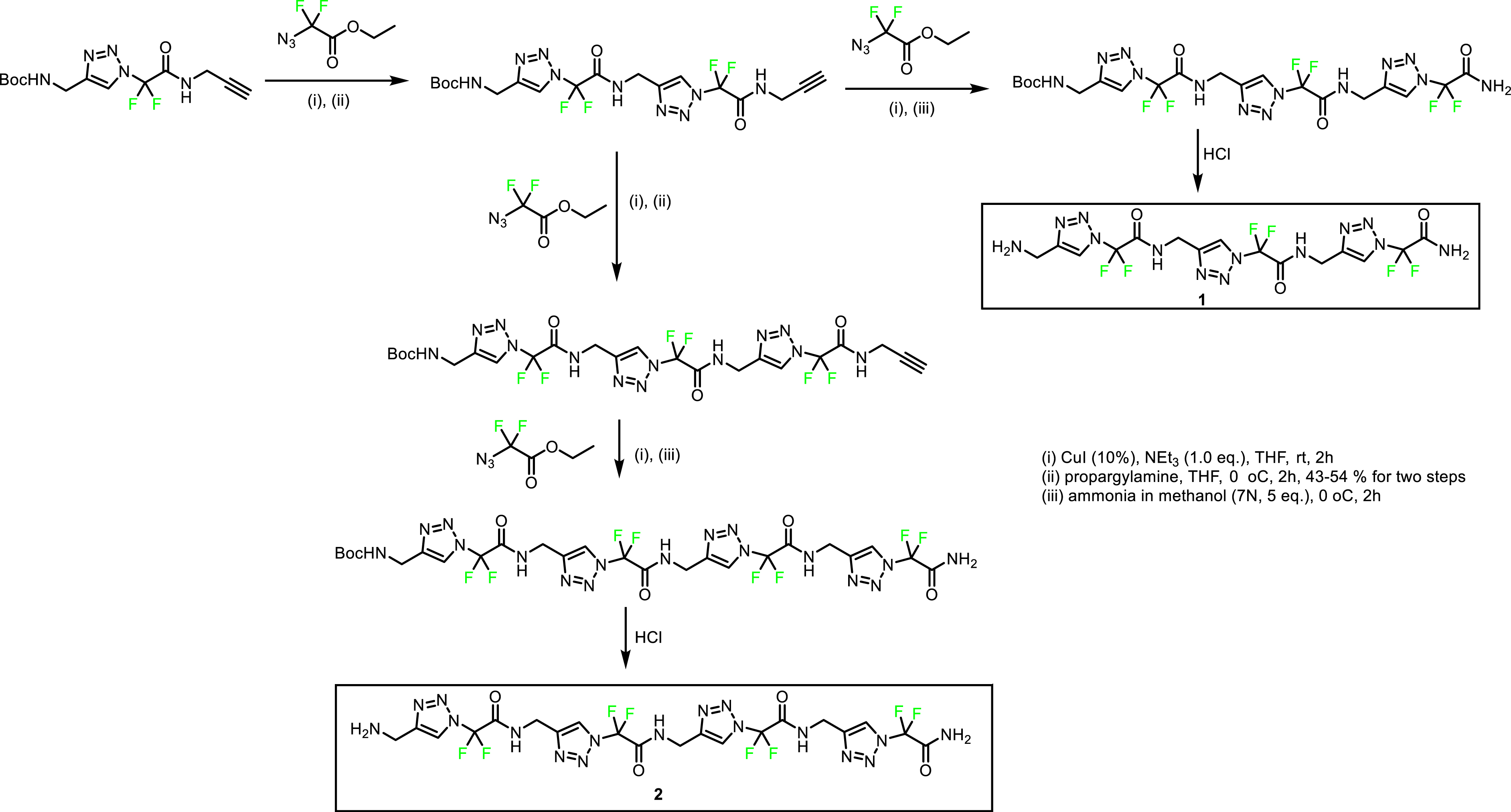
Initially, we based our attention on the successive coupling of 1,4-Tz-CF2 amino acid to prepare homotriazolamers, trimer 1 and tetramer 2, (Scheme 2, foldamers containing only triazole units) [35]. Triazolamers 1 and 2 were prepared according to Scheme 2 by carrying out sequences of click chemistry and amidation with propargylamine and ammonia, and finally Boc cleavage under acidic conditions.
4. Synthesis of heterotriazolamers
Next, we designed fluorinated heterotriazolamers by intercalating natural amino acids between the 1,4-Tz-CF2 units in order to compare the conformations of these structures with those of homotriazolamers 1 and 2. These natural amino acids were chosen according to the amyloidogenic sequence of hIAPP, N21N22F23G24A25I26L27, recognized as mainly responsible for its aggregation. The specific lateral chains of the natural amino acids should allow interaction with the amyloidogenic sequence of hIAPP, while the 1,4-Tz-CF2 units should act as β-sheet breaker elements disturbing local intermolecular interactions.
Thus, four fluorinated heterotriazolamers 3–6 were prepared for our study. Here also, foldamers were prepared first by click chemistry reactions with amino acids, then by peptide coupling reactions, and finally by Boc deprotection and acetylation (Scheme 3) [36].
Fluorinated heterotriazolamers 3–6.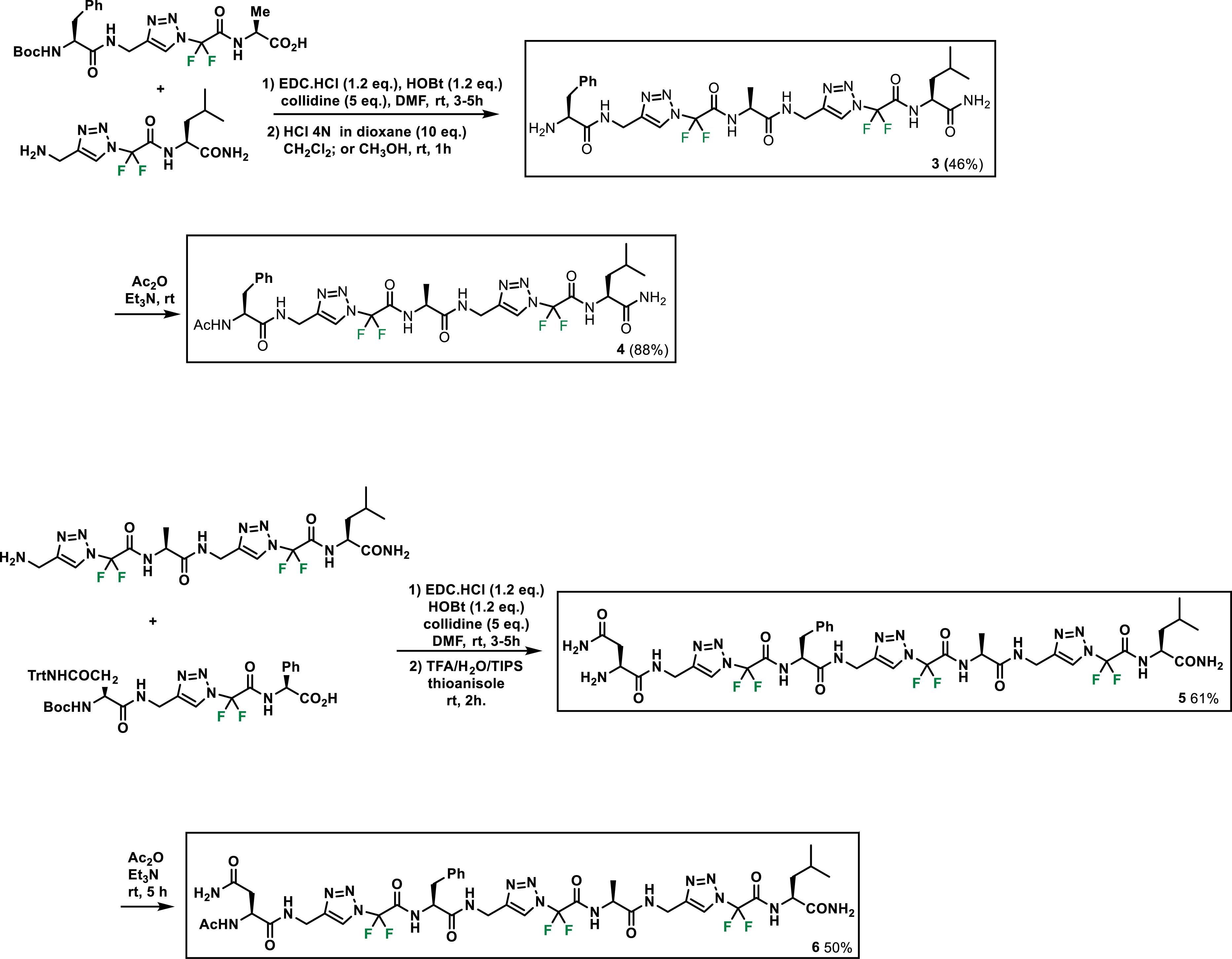
In the remainder of our study, the foldamers were subjected to conformational evaluations by NMR and molecular dynamics.
5. Conformational analysis NMR
The conformational studies of oligomers 1–6 were carried out by NMR in CD3OH, a polar solvent avoiding solubility problems in water. Many experiments such as 1D (1H,19F) and 2D (1H-1H TOCSY, 1H-1H ROESY, 1H-19F HOESY, 1H-13C HSQC, and 1H-13C HMBC) were recorded at 283 K and 313 K, respectively. The main results obtained from these different experiments are presented below.
In trimer 1 and tetramer 2, the amide NH bonds are not engaged in stable intra- (or inter-) molecular hydrogen after the calculation of the temperature coefficients. Furthermore, no long-distance correlation was observed for trimer 1 and tetramer 2. Their 2D 19F-1H HOESY analyses confirmed short-distance correlations within each N-difluoromethyl triazole unit, between the two fluorine atoms, the triazole protons, and the NH of the subsequent unit, already observed for the monomer [35]. So the rotation around the difluoromethyl group is restricted, either due to the steric hindrance caused by the two fluorine atoms or because of a stabilizing electrostatic interaction between the fluorine atoms and the neighboring protons. Furthermore, no long-range 19F-1H hetero-NOE correlations between different units were detected, supporting an extended conformation [35].
Concerning heterotriazolamers 3 and 4 with amino acids, long-range 1H-1H ROE correlations were observed, indicating folded conformations. In both 3 and 4, ROEs were observed between the CH3 of Ala-3 and the Hc of the two triazoles, TZL-2 and TZL-4. In 4, correlations between CH3 of Ala-3 and Hα of Phe-1 and between Hβ of Phe-1 and Hc of TZL-2 were also visible (Figure 2).
NMR interactions of foldamers 3 and 4 in CD3OH (1H-1H and 1H-19F NOE).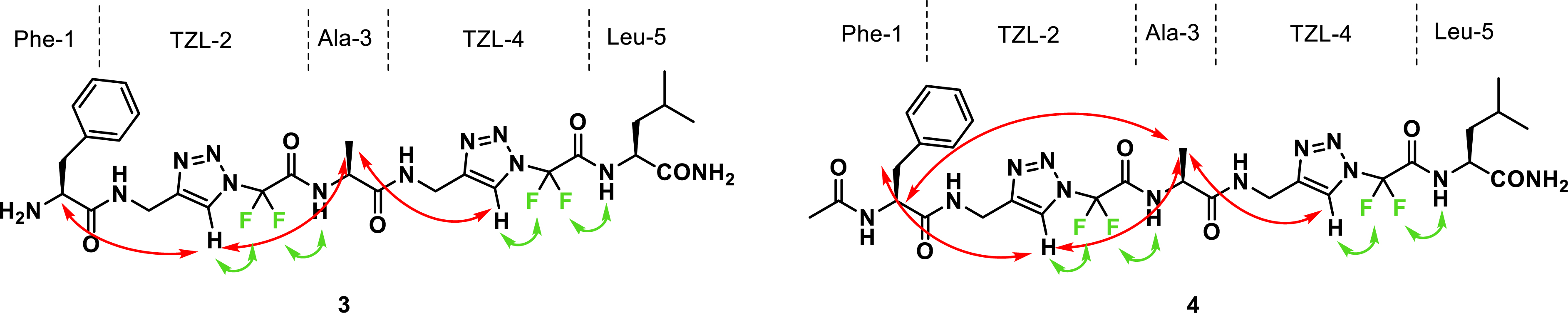
The two amino acids both at the N- and C-termini of the TZL-2 unit could become close together thanks to the flexibility of the Cα of the TZL unit. Indeed, this side of the N-difluoromethyltriazole scaffold has a higher flexibility than the side bearing the two fluorine atoms, which induces a constraining effect thanks to H–F interactions (Figure 3). In the case of the NH2-free analogue 3, one ROE was visible between Hα of Phe-1 and Hc of TZL-2. The direct rapprochement between Phe 1 and Ala-3 was not observed [36].
NMR interactions of foldamers 5 and 6.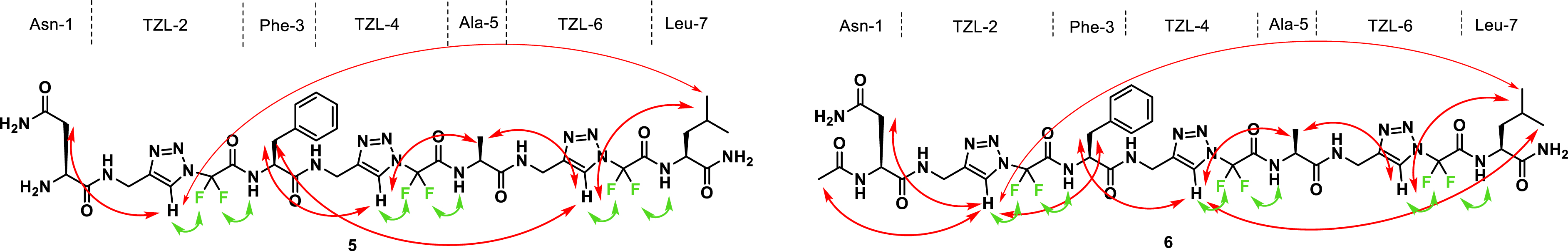
Very similar NMR data were obtained for the longer heterotriazolamers 5 and 6, confirming their folded conformations. In particular, many long-range 1H-1H ROE correlations were observed (Figure 3) (between CHβ of Asn-1 and Hc of TZL-2, CHβ of Phe-3 and Hc of TZL-2 and TZL-4 [only TZL-4 for 6], CH3 of Ala-3 and Hc of TZL-4 and TZL-6, and CHδ of Leu-7 and Hc of TZL-6). Interestingly, a very long-range 1H-1H ROE correlation was also observed between CHδ of Leu-7 and Hc of TZL-2 in both 5 and 6, indicating a close proximity between their N- and C-termini. In the 2D 19F-1H HOESY spectra of 3–6, only the short-range correlations involving in each 1,4-Tz CF2 unit the two fluorine atoms with the proton of the triazole and the NH of the successive unit were visible. This result suggests that the two fluorine atoms of each 1,4-Tz CF2 unit maintained the extended constraint at its C-terminal part but were not involved in the overall folding of the foldamers (Figure 3) [36].
6. Conformational analysis by molecular dynamics
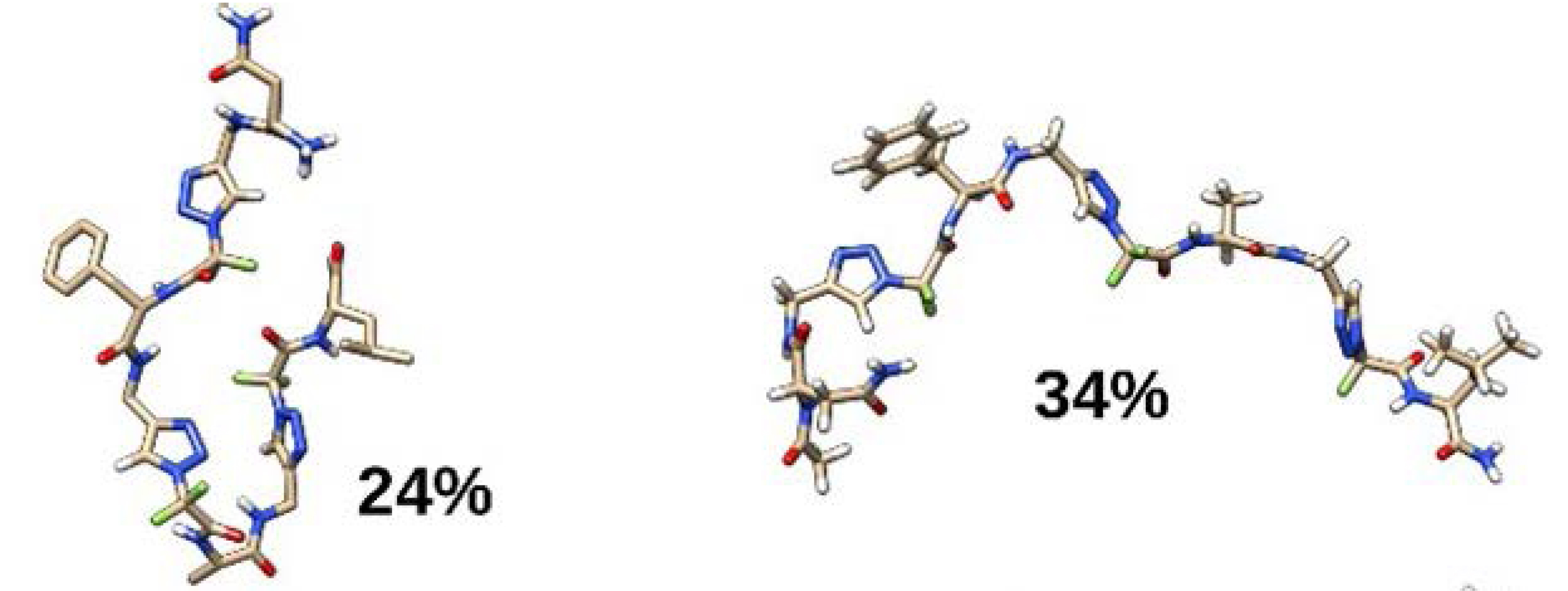
In parallel, molecular dynamic (MD) simulations of these triazolamers (1–6) were conducted in explicit methanol solvent to be as close as possible to NMR conditions. The convergence of our simulations was evaluated first by visually inspecting the time evolution of RMSD relative to the initial conformation and that of the radius of gyration (Figure 4), and by comparing distributions of the radius of gyration over time. The distributions of the radius of gyration of 1,4-Tz CF2 triazolamers 1 and 2 attain high values, indicating extended conformation (Figure 4) [35].
Representative structures of the most populated clusters of 1,4-Tz-CF2 homotriazolamers 1 (left) and 2 (right).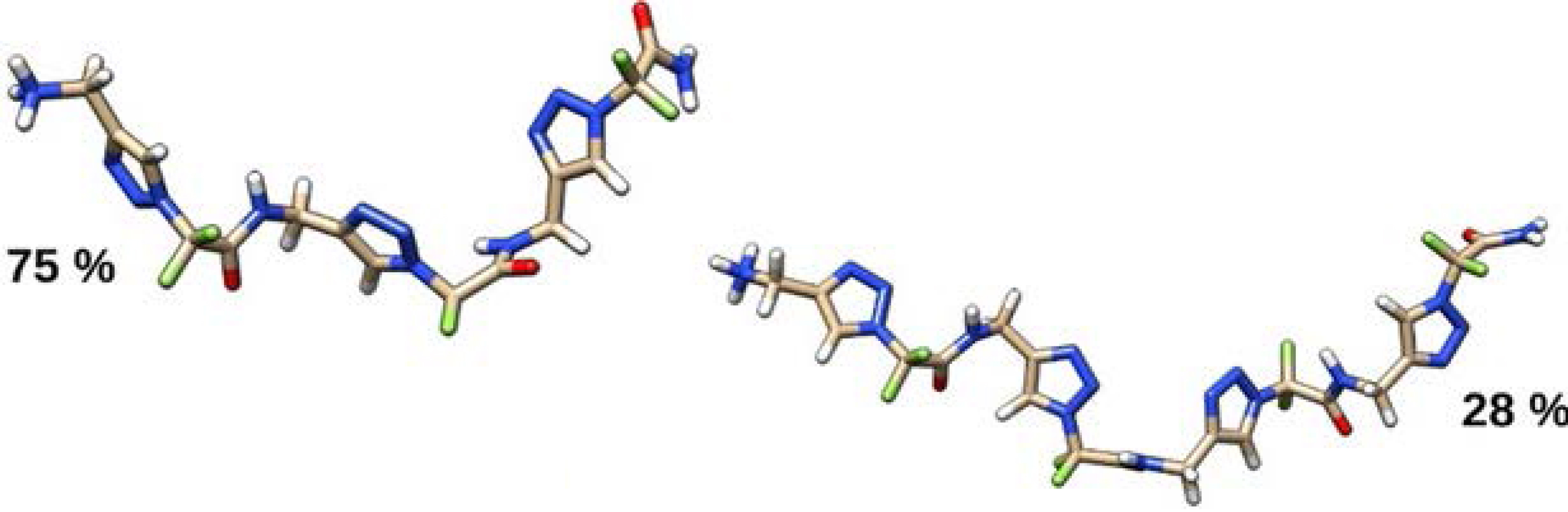
On the contrary, MD simulations of compounds 3 and 4 generated conformational ensembles in which the two rather rigid and extended segments AA-TZL2-AA and AA-TZL4-AA can rotate around the central AA residue, acting as a hinge and allowing conformational transitions between hairpin-folded segments and more extended linear structures (Figure 5) [36].
MD trajectory visualizations show that both molecules 5 and 6 are highly flexible. The representative structures of the most populated clusters of compounds 5 and 6 are displayed in Figure 6. As in compounds 3 and 4, Asn-TZL, Phe-TZL, and Ala-TZL fragments are rather rigid but can rotate around the two residues Phe and Ala, which act as bends. Thus, the transient hairpin-like structures or zigzag conformations angled at these two positions are observed, sometimes even folding into ladder-like structures similar to short multistranded β-sheets (Figure 6) [36].
As the activity of these compounds were performed in water, we compared MD simulations of compounds 4 and 6 in explicit water versus methanol. Both compounds 4 and 6 were slightly more flexible and sampled more diverse conformations than in methanol. Nevertheless, the transient hairpin-like structures of compound 4 were still significantly present in water. Notably, conformation of compound 6 appeared slightly more compact but less structured in water than in methanol.
7. Activity on hIAPP Fibrilization by ThT-fluorescence experiments
The ability of foldamers 1–6 to interact with hIAPP fibril formation was first investigated using the in vitro Thioflavin T (ThT) fluorescence assay. We only report here triazolamers that showed some specific effect and activity on amyloid, compared to the natural peptides FGAIL-NH2 and NNFGAIL-NH2. The aggregation curves of hIAPP in the presence of homotriazolamer 1 at 1/hIAPP ratio 10/1 (Figure 7) showed a shorter t1/2 and a higher fluorescence plateau than for the control experiment, indicating that compound 1 is capable of accelerating but also increasing the aggregation of hIAPP [36].
Representative curves of ThT fluorescence of one assay over time showing hIAPP control aggregation (5 μM) in the absence (red curve) and in the presence of compounds 1 and 4 at compound/hIAPP ratios of 10/1 (purple curves) and 1/1 (green curves). The control curves are represented by orange lines and the buffer by gray. Solid lines correspond to curves fitted with a Boltzmann sigmoidal model.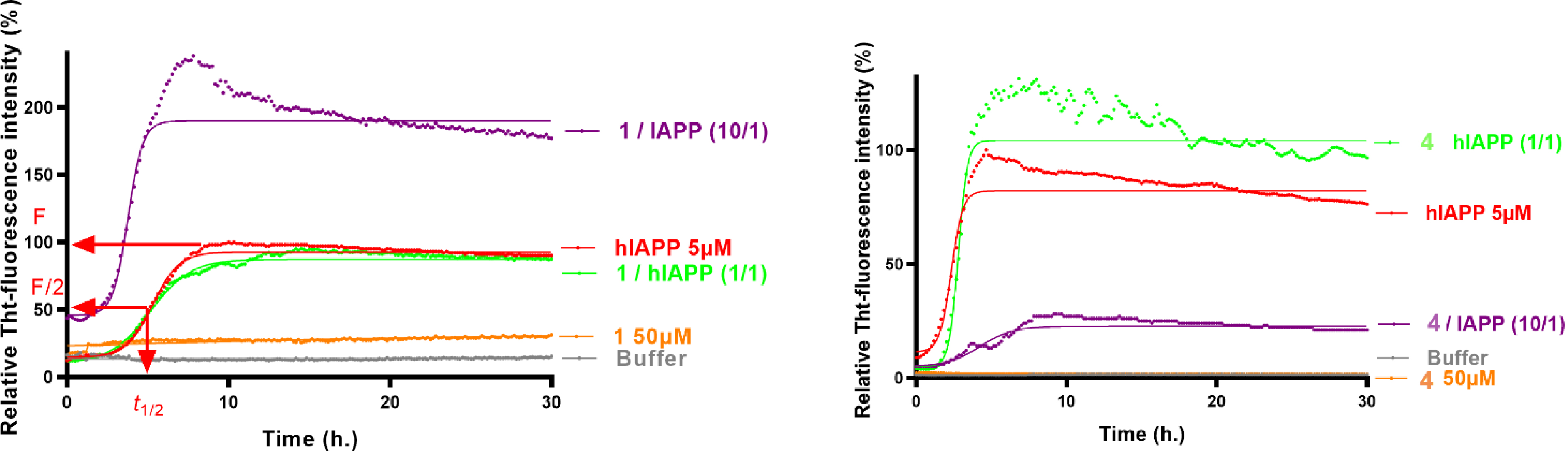
On the contrary, the inhibitory activity of 4 was dramatically increased compared to that of the natural peptide parent FGAIL at 4/hIAPP ratio 10/1 (Figure 7), both on the fibrilization kinetics (Δt1/2 = +112% for 4 vs no effect for FGAIL-NH2) and on the fluorescence plateau (ΔF = 80% for 4 vs = 36% for FGAIL-NH2). The activity was not retained at the lower ratio of 1/1.
8. Interaction of homotriazolamer 1 and heterotriazolamer 4 with hIAPP by NMR
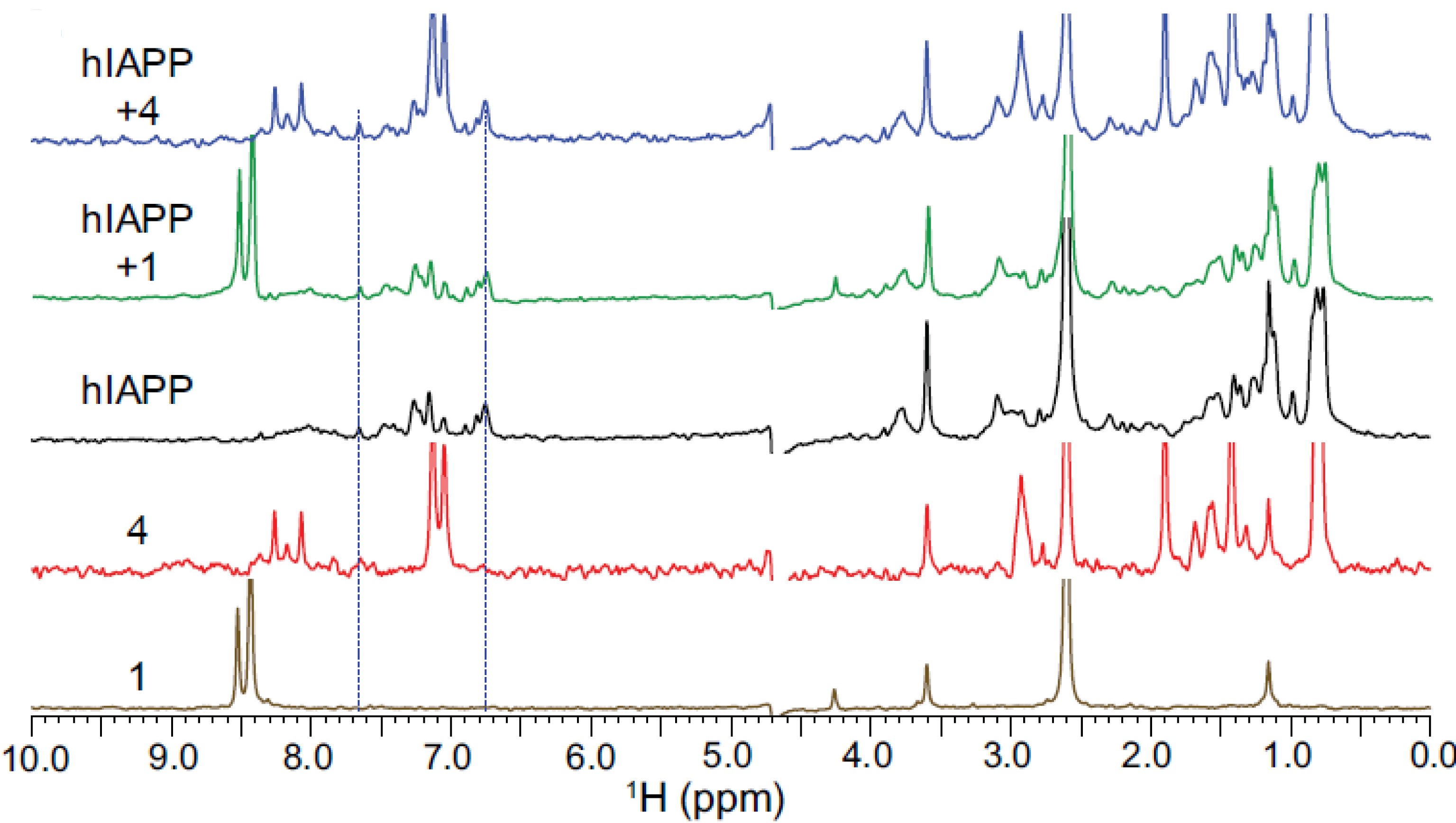
The effect of compounds 1 and 4 on hIAPP was first studied using proton NMR experiments (Figure 8). In particular, for 4, interactions with sequences directly involved in the hIAPP aggregation process (residues 10–17 and 25–28) were observed. At 0.5 h, a line broadening was observed in the presence of 1 or 4 (green and blue curves, respectively), which could be due to their interaction with monomeric hIAPP or to hIAPP aggregation.
The interaction of compounds 1 and 4 with hIAPP was then investigated using 19F NMR experiments (Figure 9). For the achiral homotriazolamer 1, which contains three CF2 groups, the 19F NMR signals appear as three very close singlets around 88 ppm. When 1 was mixed with 50 μM hIAPP, a change in signal intensities around 88 ppm was observed (green spectrum compared to the orange spectrum of the compound alone; see Figure 9). In the case of compound 4, mixing it with hIAPP resulted in a roughly 40% reduction in the 19F signal intensity in the 86–88 ppm region compared to 4 without hIAPP (blue spectrum compared to red spectrum; see Figure 9).
19F NMR spectra of 1 and 4.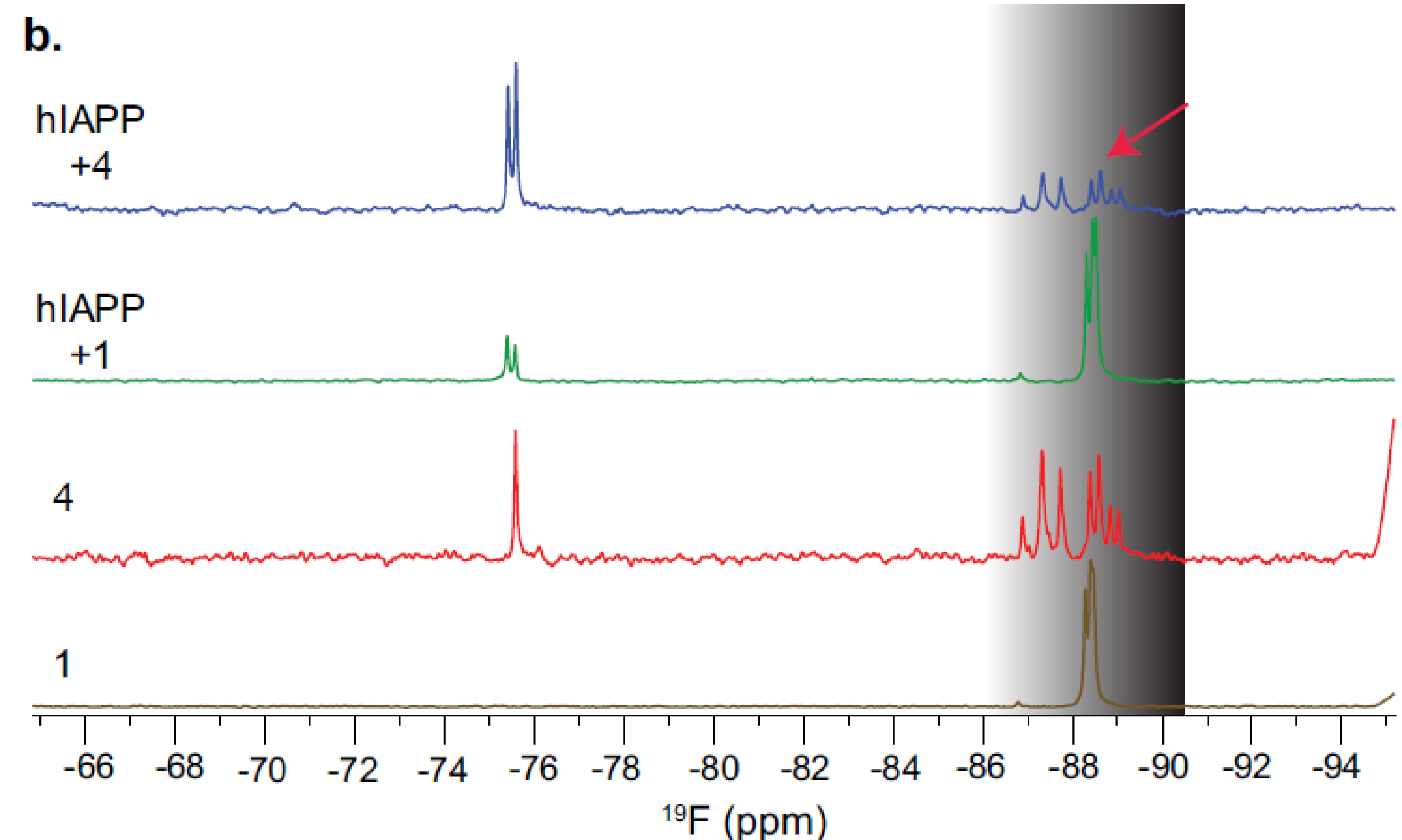
The observed differences in signal intensities can be attributed to the presence of two distinct populations of the compounds in the mixture: those bound to hIAPP and those not bound to hIAPP in a mixture containing a 5 molar excess of compounds, specially in the case of heterotriazolamer 4.
9. Effect on hIAPP oligomerization by native and ion mobility mass spectrometry experiments
Mass spectrometry (MS) has been used as a valuable technique to identify oligomeric species formed during the early oligomerization process of amyloid peptides and proteins. MS experiments revealed differences between the two foldamers 1 and 4, in particular regarding the nature, number, and shape of noncovalent complexes of foldamers with hIAPP monomers/oligomers formed. Foldamer 1, which accelerates fiber formation in ThT experiments, interacts slightly with the hIAPP monomer while the noncovalent complexes hIAPP/1 and hIAPP oligomers disappear very quickly [36].
On the contrary, foldamer 4, which is a retarder of the fibrilization process in ThT assays, forms numerous noncovalent complexes almost exclusively with the hIAPP monomer, which are more compact and stable over time. We can therefore hypothesize that foldamer 4 interacts more strongly with hIAPP monomers, sequestering them and favoring a compact conformer that may not be prone to aggregation.
10. Conclusion
We demonstrated that fluorinated triazolamers based on aminomethyl-1,4-triazolyl-difluoroacetic acid (1,4-Tz-CF2) are readily available synthetically. Experimental and computational conformational studies showed that homotriazolamers adopt extended or worm-like chain structures, while heterotriazolamers are structured more like hairpins. Some of the triazolamers have given rise to very interesting but opposite activities on hIAPP, with one homotriazolamer accelerating and one heterotriazolamer inhibiting hIAPP aggregation.
This study highlights the potential of hitherto unknown fluorinated peptidomimetic foldamers as modulators of amyloid aggregation as well as for wider implications in medicinal chemistry. Our findings affirm the value of fluorine “labeling” in investigating biological events, such as ligand–biomolecule interaction structures via 19F NMR spectroscopy. In addition, this work paves the way for utilizing fluorinated foldamers in 19F MRI medical imaging.
The interest of substituting the methylene of the aminomethyl-1,4-triazolyl-difluoroacetic acid to mimic other AA-difluoroGly dipeptide to increase the affinity towards other target proteins will be soon provided.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.
Acknowledgments
The authors gratefully acknowledge all the students and collaborators who have contributed to this work. They thank the CNRS and the French Ministry of Education and Research for their financial support. They also warmly thank the French Fluorine Network (GIS-FLUOR) for its unconditional support.





 CC-BY 4.0
CC-BY 4.0