1 Introduction
Creating life in a test tube has been a powerful source of inspiration for fiction writers and journalists, but has also always fascinated scientists themselves. In the 19th century, the first chemical synthesis of an organic molecule, urea, was considered to be a landmark in the fight of science against vitalism. In the 1920s, the synthesis of DNA in a test tube had a similar impact [1], probably explaining why Arthur Kornberg, who discovered the first DNA polymerase in 1958, won the Nobel Prize in 1960, two years before Watson and Crick received the prize for their double-helical model of DNA [2]. The complete in vitro synthesis of the DNA genome of the bacteriovirus ΦX174 in 1967 in the laboratory of Arthur Kornberg [3] has been another step in the artificial synthesis of life, widely celebrated at that time by the media. However, everybody has agreed that a genome by itself is not a living organism.
Although the artificial synthesis of proteins does not make much sense, it should be theoretically possible in the near future to synthesize a completely artificial viral particle by combining in vitro synthesized DNA or RNA with in vitro synthesized capsid proteins using only tools from synthetic chemistry. This would be certainly acclaimed by some (especially journalists) as the synthesis of life, and discredited by others, mostly those scientists who argue that viruses are not living entities. In this article, I will use this question as a starting point to present recent developments around the concept of viruses: should we confuse viruses and virions? And, if not, what is a virus? I will discuss recent suggestions to identify the virus with viral factories or else to consider them as atypical “cellular organisms”. I will then introduce the concept of virocell, in which the infected cell is no longer viewed as a near-dead cell, but as an active living being, the virus itself. The virocell concept emphasizes the unique capacity of viruses to manipulate biological objects (molecular and cellular) and create new ones. This suggests to take into account more seriously viruses as possible source of new tools and new manipulators (beside ourselves) in synthetic biology.
2 Virions are not living… but are also not viruses
2.1 The traditional view confuses viruses and virions
Are viruses living? Many biologists have answered no to this question because they assume that the viral particle, the virion, and the virus are one and the same thing. This immediately implies that viruses are not living organisms, since, traditionally, all living organisms are supposed to be made of cells. The influence of the cellular theory: Omniae cellula e cellula, and the concept of “viruses as virions” thus mainly explain why viruses have been so often expelled from the realm of life. Virions indeed are not living organisms, they are inert until they encounter cells (for an exception, see [4]). As a matter of fact, virions have no active metabolism, they cannot produce energy, and some of them can be crystallized. However, the virion state only represents one form of the virus life cycle. In a classical review paper entitled “viruses and genes”, Jacob and Wollman wrote in 1961 that “a virus may exist in three states: the extracellular infectious state, the vegetative state of autonomous replication and finally the proviral state” [5]. The term “vegetative state” has been probably misleading, since it has a negative connotation (at least in French) meaning that a person in such state is still alive but cannot think anymore (thus is not really alive according to our subjective homo sapiens criteria). Indeed, in agreement with the traditional view on the nature of viruses, Jacob and Wollman finally defined the virus as “a genetic element enclosed in a protein coat”, i.e., a virion [5].
Our long time assimilation of the virion to the virus can be easily explained. We need to associate a precise shape to an organism and the virion fulfils this requirement. Amazingly, we indeed distinguish in the virion of bacterioviruses such as T4, a head and a tail, as if they were mini animals. In the last years, a sphere decorated with harmful spikes is becoming the common representation of viruses on TV. In contrast, the “vegetative state of autonomous replication” was named the “eclipse phase” and was, of course, refractory to any clear-cut representation, not the least anthropocentric ones. To my knowledge, the only biologist who, for a long time, has proposed an alternative view of viruses is Claudiu Bandea, who wrote in 1983 that “The living phase of the virus is the intracellular phase of its life cycle” or else the “in the vegetative phase, the virus shows the major physiological properties of other organisms: metabolism, growth, and reproduction. Therefore, life is an effective presence” [6]. However, these remarks went mostly unnoticed and one had to wait 2006 for a direct attack against the classical view equating viruses and virions (for recent papers by Claudiu Bandea, discussing the origin, role and nature of viruses, see [7]).
2.2 A challenge: viruses as viral factories
Following the discovery of the giant Mimivirus in the laboratory of Didier Raoult [8], Jean-Michel Claverie, who was involved in the sequencing and analysis of the Mimivirus genome, wrote in 2006, “This traditional view… the virion is the virus… might, however, be a case of ‘when the finger points to the stars, the fool looks at the finger’. Rather than comparing a parasitic cell to the virus particle, I believe we should compare it to the ‘virus factory’, and also “Interpreting the virion particle as ‘the virus’, is very much like looking at a spermatozoid and calling it a human” [9].
Mimivirus infects the protist Entamoeba haemolytica, a large eucaryotic microbe [7]. The viral particle of Mimivirus is as big as a mycoplasma (a small bacterium) and its genome (1.3 Mb, encoding probably around 1000 proteins) is more than two times larger than the genome of a small bacterium [8]. The virions of Mimivirus are produced at the outer surface of spherical structures, the viral factories, which are as big as the nucleus of the amoeba and can be seen by light microscopy [10]. Viral factories have well defined borders, allowing then to give a physical reality to the “vegetative state of autonomous replication” which, otherwise, remains a paper scheme. The viral infection led to a reorganization of the cellular metabolism with the active participation of viral encoded proteins, as illustrated by the presence of many “metabolic genes” in the Mimivirus genome. The viral factory thus looks like an intracellular parasitic organism endowed with metabolic activity. Finally, the viral factory of the Mamavirus, a close relative of Mimivirus, can be infected by a small virus, Sputnik [11]. This infection reduces the production of Mimivirus and leads to the formation of abnormal virions. Viral factories thus exhibit all hallmarks of a living organism, reproduction, metabolic activity, they can even get sick [12]!
2.3 What about viruses without viral factories?
The viral factory was not a new concept by itself. All viruses infecting eukaryotes, including RNA viruses, manipulate the intracellular membrane network to produce viral factories [13,14]. One can thus wonder why the concept of “viruses as viral factories” was not proposed earlier. This is probably because our present view on viruses mainly originated from studies of the “phage group” in the 1950s and 1960s. The study of bacteriophages does not led to the discovery of viral factories inside the bacterial cell. Bacteria being considered as “bag of enzymes”, it was largely believed that all viral components were indeed simply dispersed in bacterial cytoplasm during the eclipse phase.
The question in fact remains, are the viral factories of bacteriophages too small to be visible? Did they exist at all? If eukaryotic viral factories are characterized by the recruitment of endoplasmic reticulum membranes, it might be that viral factories do not exist in Bacteria and Archaea because most of them have no such system of intracellular membranes (for an exception in the bacterial domain, see the case of Planctomycetes [15]).
I recently discovered an early study by thin-section electron microscopy of Escherichia coli cells infected by T-even bacteriovirus (T2, T4) that directly address the problem discussed here [16,17]. In this work, Simon has observed that the production of virion head and tails both occur independently at the inner surface of the cytoplasmic membrane, thus mimicking the building of the virions of eukaryotic viruses at the outer surfaces of viral factories membranes. Once built at the membrane level, the T4 heads leave the membrane to be filled with DNA within the cytoplasm. The full head is then attached to the tail still bond to the membrane (Fig. 1). Interestingly, electron dense structures look like mini viral factories that can be seen inside the infected cell at the onset of the “eclipse” phase. However, these structures correspond in fact to the accumulation of capsid protein as precursors of head formation [17]. Similarly, we failed to detect bona fide viral factories in studying by thin-section electron microscopy cells of the archaeon Sulfolobus islandicus infected by the virus SIRV [18]. We could nevertheless detect unusual large pyramidal structures, located at the cell envelope, that seem to be used to perforate the membrane and S-layer, allowing bunches of linear virions to escape from the cell.
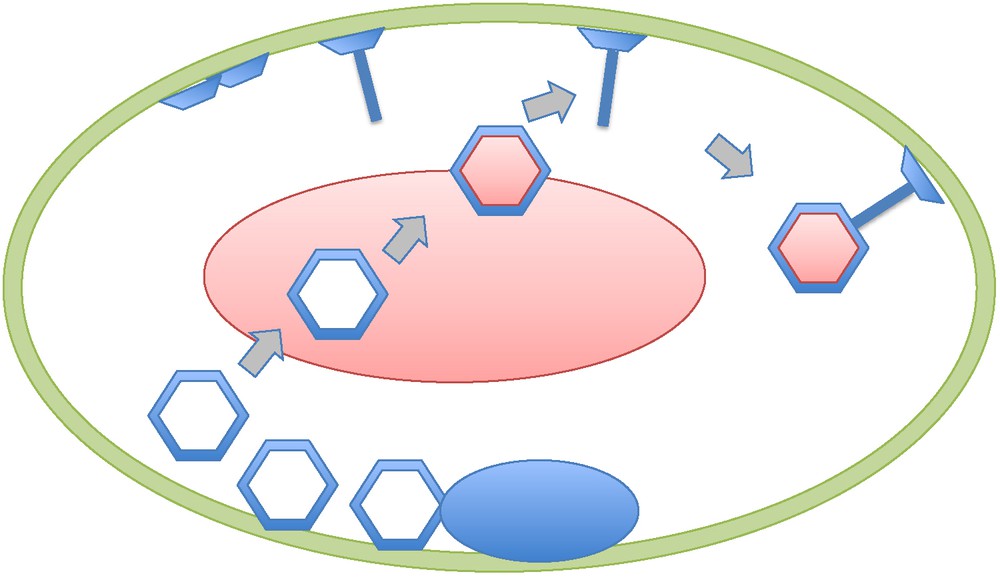
Schematic representation of a T4 virocell (adapted from [16,17,22]). The capsid proteins accumulate first as condensed material looking-like a viral factory (black oval). Heads and tails (with plates) are made independently at the level of the former Escherichia coli membrane which has been modified by the presence of T4 proteins. Once completed, the empty heads migrate toward the centre of the virocell to be filled with T4 DNA (grey oval). The bacterial DNA has been completely destroyed and the only “cellular” DNA present is the viral DNA. After DNA filling, the heads are fixed to the tails bound to the virocell membrane.
3 The virocell concept
3.1 The infected cell is the viral organism
We are now facing the following problem: if viral factories, as described for viruses infecting eukaryotic cells, do not exist in the case of viruses infecting archaea or bacteria, the concept of “viruses as viral factories” cannot be a general concept to replace the traditional one of “viruses as virions”. One way to get out of this conundrum is to refer to André Lwoff who wrote in his famous paper on the definition of viruses that “the virus transforms the cell into a viral factory” [19]. Considering that infected cells in fact produce virions and not viruses, this sentence can be translated, “the virus transforms the cell into a virion factory”. Now, if we define viruses as organisms producing virion (capsid-encoding organisms), we can write: “the virus transforms the cell into a virus”. This leads David Prangishvili and myself to suggest that the infected cell is the real viral organism [20,21]. This idea is best illustrated by the behaviour of viruses that completely degrade the chromosome of the infected cell, as it is the case for T4 (Fig. 1) or for the archaeal virus SIRV 2 [18]. The only genetic material in such an infected cell is therefore that of the virus itself. The cells are now completely remodeled in order to produce virions, its proteome being composed by a mixture of old proteins encoded by the now extinct cellular genome and new proteins encoded by the active viral genomes. As a consequence, all syntheses (and metabolic activities in general) that occur inside such cell are now dictated by the viral genome. For instance, T4, as many other viruses, produce many proteins that can be integrated into the membrane or modify its properties (e.g., lipid composition) for its own purpose [22]. In addition to bearing a new genome, the infected cells are thus surrounded now by a “new” membrane. The production of pyramidal structures to create holes in the membrane of infected archaeon is another spectacular example of cell remodeling by viruses [18]. In such cases, it is difficult to consider the infected cell as a bacterium or an archaeon anymore, since it has no longer the corresponding genome, but a virus expressing itself with a cellular shape.
I suggest here to call such cells “virocells” (Fig. 2). In this framework, viruses are cellular organisms, but of a very atypical kind. To paraphrase Jacque Monod in “Chance and necessity” [23], whereas the dream of a normal cell is to produce two cells, the dream of a virocell is to produce hundred or more new virocells through the dissemination of virions (Fig. 2).
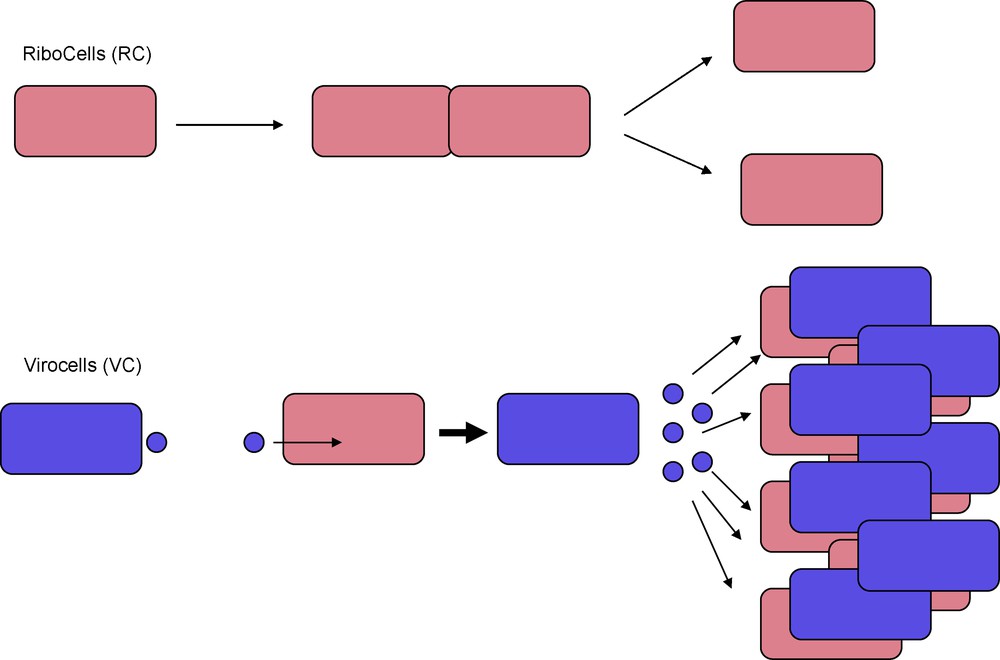
Ribocells and Virocells. Ribosomes encoding organisms (30) are made of ribocells that reproduce by cell division. Capsid encoding organisms (30) are made of virocells that are obligate parasites of ribocells. Virocells reproduce by producing virions that carry their genetic material to infect new ribocells. Unlike ribocells, virocells cannot reproduce autonomously, but their propagation mechanism is much more effective.
3.2 Some implications of the virocell concept
The concept of virocell has important consequences. First of all, the suggestion to consider viruses as cellular organisms implies that viruses are definitely living and should be included in the definition of life (see [24], for a discussion on the definition of life). Unlike the recent proposal of Bandea to consider viruses as “molecular organisms” [7], The virocell concept thus reconciles the idea that viruses are living with the classical view that all living organisms are made of cells. This concept then clarifies the initial question raised in this paper, making obvious that chemical synthesis of a virion will not be sufficient to claim that we have synthesized life in a test tube. The virocell concept also reminds us that viruses could not have appeared before cells, since they are themselves cellular by essence. This concept is therefore not compatible with “virus first” theories for the origin of life, in agreement with the widely spread view that formation of open thermodynamic systems with well defined boundaries (cell-like structures) has been a critical step (possibly the first one) in the origin and evolution of life [25]. The question of the origin of viruses then becomes the question of the origin of virions as a specific mechanism of gene dissemination in the RNA/protein world (for the discussion of various scenarios for the origin of viruses in such cellular context, see [6,20,26]).
Incidentally, the virocell concept led to a reappraisal of the abundance of viruses on our planet. It is often stated that viruses are more abundant than cells. For instance, a current statement is that “viruses are ten times more abundant than bacteria in the ocean”. This sentence should be read now “virions are ten times more abundant than cells in the ocean”. To paraphrase Claverie, counting virions as viruses is equivalent to count the number of fish sperm cells in the ocean to estimate the number of fishes!! In the framework of the virocell concept, viruses cannot be more abundant than cells, but a fraction of cells on our planet. For example, it has been estimated that on the average, about 20% of marine heterotrophic bacteria are infected by viruses [27]. One can conclude from such estimate that 80% of marine bacteria are real bacteria, whereas 20% are virocells, corresponding to living bacterioviruses.
3.3 Virocells and ribocells
Although the virocell concepts put the emphasis on the active living stage of the virus life cycle, it does not contradict the idea to define viral lineages based on the structure of virion proteins (the virus self, [28,29]), since the virion and its capsid remains the hallmark of viruses (allowing them to be distinguish from plasmids, or else viroids). Similarly, the virocell concept does not contradict the definition of viruses as capsid-encoding organisms [30] since the specific characteristic of virocells is to produce virions (most often with capsid). The virocell thus represents the cellular phase of the capsid-encoding organism. To achieve its objective, the virocell need to infect the cell of a ribosome-encoding organism. I suggest here to call “ribocells” the cells encoding ribosomes to distinguish them from virocells (Fig. 2). The virocells and ribocells are two very different entities, at least in our modern world, they use completely different mechanisms to disseminate their genes (cell division for ribocells versus production of virions and for virocells) and only ribocells can live without virocells, whereas the opposite is not true. To avoid confusion, will for instance reserved the term cellular genome for the genome of ribocells.
3.4 Virocells and the origin of the eukaryotic nucleus
The concept of virocells raises new amazing theoretical questions with evolutionary implications. For instance, in the case of viruses infecting eukaryotes, if we consider that the infected cell is a virocell, what is the viral factory? In fact, it is tempting to assimilate the viral factory to the nucleus of the virocell. Interestingly, several authors have proposed an evolutionary link between giant DNA viruses and the eukaryotic nucleus [31,32]. They suggested that the eukaryotic nucleus originated from a large DNA virus (NCLDV-like) that infected an archaeon (the viral eukaryogenesis hypothesis). This idea was not taken seriously by most evolutionists because, confusing the virus and the virion, they could not imagine how a virion could become a nucleus and found this hypothesis ridiculous. However, the transition from a Mimivirus-like viral factory to a nucleus is much less difficult to imagine. Amazingly, the viral factories of Vaccinia viruses are indeed called “mininuclei” by virologists considering the striking similarity between these two structures [33]. Viral factories and eukaryotic nuclei are both replicating in the cytoplasm and their membrane is built from those of the endoplasmic reticulum.
Eukaryotic chromosomes are at first glance very different from any known viral chromosome structures. However, the telomers and centromers most likely originated from retroviruses (see below). Since NCLDV with a linear chromosome can harbour integrated retroviruses in their genomes [34], it is not so difficult to imagine how an ancestral viral chromosome might have been remodeled into a modern eukaryotic one in the proto-eukaryotic lineage. If the eukaryotic nucleus indeed evolved from one or several viral factories of large DNA viruses, it is, however, unlikely that these NCLDV-like virus infected a modern-looking archaea, such as a methanogen, as suggested by Takemura and Bell [31,32]. These viruses probably infected already large ribocells with extensive intracellular membrane networks that could be more cautiously called proto-eukaryotic cells [35]. Alternatively to the viral eukaryogenesis hypothesis, Bandea suggested that the nucleus originated as a protection device against viruses (anti-fusion mechanism) [36]. Since one of the functions of viral factories is probably to protect the viral genome from cell nucleases, it is indeed possible that early eucaryotic cells learned how to protect their genomes against viral nuclease attack by building viral-like factories (nucleus ancestors) using viral tools [21].
3.5 Two organisms in the same cell
Finally, the virocell concept raises fascinating questions regarding the proviral state, a situation in which two organisms, the ribosome encoding organism and the virus cohabit in the same RiboVirocell (Fig. 3). In that case, it is tempting to conceptually reduce the organisms to their genomes, telling that the two genomes coexist in the same cell. However, an organism cannot be reduced to its genome, especially because the evolution of genomes and organisms are two different things [37].
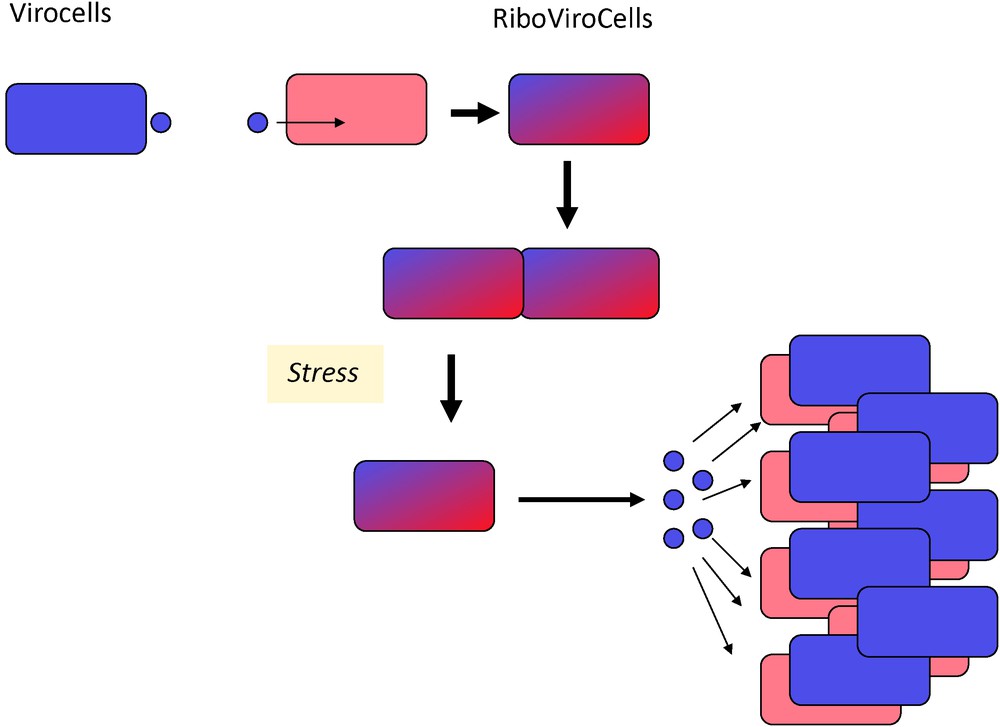
RiboVirocells. Infection of a ribocell by a virocell can produce a lysogenic state in which the cellular and the viral genomes coexist in the same cell, the RiboVirocell. In this state, two organisms (for instance a bacterium and a bacteriovirus) are living in the same cellular structure in a delicate equilibrium. Stress treatment usually shifts the equilibrium in favour of the virocell that initiate its reproductive cycle and kills the ribocell.
In the “proviral state”, the two organisms can establish various levels of coexistence for very long periods. The virocell can be physically reduced to its genome (usually integrated into the genome of the ribocell) and a few expressed proteins, or to be partly active, producing virions without killing the host, as in the case of the bacteriovirus M13. Coevolution of the two organisms in this persistent state will have important consequence on their history, as they will adapt to each other [38,39]. In many cases, the ribocell will succeed to definitely inactivate the virus in such way that only one of the two organisms (the ribocell) survives. However, it usually still contains relics of the infection and these relics (viral proteins) can become essential to the survival of the ribocell, providing crucial new functions.
4 Viruses as masters of synthetic biology
4.1 The origin of viral genes
Importantly, the concept of virocell emphasizes the capacity of viruses to manipulate ribocell syntheses for their own purpose. They perform this task mainly by creating new genes to manipulate those of the ribocells or to invent new structures. However, this is not recognized by many evolutionists who still deny the existence of bona fide “viral genes”, i.e., a gene that originated in viral lineages. In their views, all viral genes should have ultimately a (modern) cellular origin and viruses are defined as pickpocket or else robbers of cellular genes [40]. In my opinion, the difficulty to imagine that new genes can originate in viral lineages came again from the confusion between the virus and the virion. Since virions are inert, many authors finally deny or grossly underestimate the synthetic capacity of viruses, their ability to create novelties. The virocell concept solves this conceptual problem. It is well known in fact that new genes appear continuously in viral lineages by various mechanisms during replication and recombination of viral genomes. This can occur by classical mutagenesis (point mutation, insertion or deletion) but also by extensive recombination, frameshift and creation of overlapping genes.
The mechanisms for the creation of new genes are similar in ribocells or virocells, but some mechanisms are probably favoured in cellular versus viral genomes and vice versa. In particular, the genome size limitation dictated by the packaging capacity of the virion and the tendency for natural selection to favour rapid replication and genome reduction in parasitic organisms, probably explain why viral genomes contain a higher proportion of overlapping genes than genomes of ribocells. The creation of new genes overlapping the sequence of an old one (overprinting) seems to be a process especially frequent in viruses, especially RNA viruses [41,42]. In contrast, creation of new genes by gene or genome duplication is probably less frequent in viral than in cellular genomes, although such an event could be compensated by concurrent mutation in capsid genes increasing the size of the virion [42]. Genes and genome duplication have been indeed well documented in the Mimivirus genome [8], but also in bacterioviruses, such as T4 [22].
4.2 Virocells and the expending virome
The creation of new genes in viral lineages is driven by the continuous selection of novel functions to counteract the protection of the host and to manipulate the host biology [43]. This leads in particular to the production of many small proteins that interact with ribocell proteins to modify their function in favour of the virus. This probably explains the plethora of small genes encoding uncharacterised proteins in viral genomes. As the consequence of this continuous gene creation that has started even before the origin of modern cells (the descendant of Last Universal Cellular Ancestor (LUCA), see below) the genetic diversity of viruses is now much greater than those of ribocells, something becoming more and more apparent with recent metagenomic studies focusing on “viromes” [44,45].
Only a fraction of the genes created in viral genomes finally end up in cellular genomes. This only occurs if the viral genome is integrated into the genome of a ribocell and if the novel proteins turned out to be useful for the ribocell. However, considering the huge number of viral genes and the continuous invasion of cellular genomes by viruses and viral derived mobile elements (plasmids, transposons, integrons, patogenicity islands), it is not surprising to find that the genomes of modern cells are full of genes of viral origin. For instance, it has been recently estimated, using stringent criteria that, on average, 13% of genes in archaeal and bacterial genomes have been recently introduced by viruses or related mobile elements [46]. Importantly, this work has shown that the majority of ORFans in archaeal and bacterial genomes are located in viral-related elements, in agreement with the idea that most ORFans in ribocell genomes originated from a huge reservoir of viral genes. The number of genes that originally were created in viruses can be much higher in the genomes of an eukaryotic organism with less size constraints. Hence, it has been claimed that up to 40% of the human genome has originated from retroviruses [47] and this figure can be even higher if one takes into account repeated sequences that seem to be related to various families of retroelements. Indeed, it has been recently shown that transposable elements, that should have ultimately originated in viruses, constitute 85% of the maize genome [48].
4.3 Viral experiments and the origin of evolutionary novelties
Viruses are very ancient, as indicated by the existence of viruses with homologous structural and replication proteins in the three domains of life [26,28,29,49–52]. They most likely predated the divergence of the three cellular domains, Archaea, Bacteria and Eukarya [49,51]. Viruses are therefore older than modern ribocells (the descendants of LUCA) and probably originated in the protein/RNA world by reduction of parasitic RNA cells and/or when the invention of virions allow some replicons to bypass the process of cell division to disseminate more rapidly their genes [21,26]. Alternatively, they possibly originated from even older protoviruses, i.e., infectious lipid vesicles containing RNA [53]. As masters in synthetic biology, viruses should have therefore played a decisive role in the evolution of life very early on by promoting the emergence of new defensive or aggressive mechanisms that were later on recruited by ribocells for their own purpose. For instance, Brussow [54] suggested that pathogenic bacteria recruited from viruses virulence mechanisms to kill their protists predators. These counter-attack mechanisms became later on pathogenicity islands when bacteria learn how to infect eukaryotic cells.
An intriguing possibility is that some protection mechanisms that originated in the framework of the fight between ribocells and viruses have been major actors in the transition from the RNA to the DNA world [35,49,55]. In that scenario, ribonucleotide reductases, thymidylate synthases, and reverse transcriptases first originated in virocells to produce new forms of viral genomes resistant to the RNAse or anti-RNA mechanisms produced by ancient ribocells. Again, it has been difficult for some biologists to understand what does it mean, how does a virus “invent” DNA? However, it has been no more (nor less) difficult for a RNR gene, or a thymidylate synthase gene to appear in a viral genome than in a cellular genome, since, as we saw previously, new genes can originate in both types of genomes from the same types of mechanisms.
In the “viral origin of DNA” scenario, the synthetic ability of viruses were also at the origin of most of the enzymes that deal with DNA for replication, repair and recombination (3R proteins), explaining why 3R proteins are more diverse in the viral world compared to the (ribo)cellular world. The hypothesis is that only a fraction of the 3R proteins created in virocells were later on recruited by lineages of modern DNA ribocells, possibly several times independently and at different periods of evolution [56]. Ancient DNA viruses might have indeed shaped the structure and replication mechanisms of modern ribocell genomes, as suggested by the similarities between archaeal and bacterial genomes with those of circular double-stranded DNA viruses, or else by the similarities between the linear chromosomes of eukaryotes and those of some NCLDV [35]. A direct and decisive role of viruses in the emergence of the unique eukaryotic chromosome is indeed suggested by the homology between reverse transcriptase and telomerase [57], by the telomer-like structure of Penelope retroelements [58] or else by the role of retroelements in the formation of centromers [59].
Finally, several authors have suggested that the formation of the three lineages of modern ribocells themselves (Archaea, Bacteria and Eukarya) has been driven by process involving viruses [21,35,36,53]. The origin of some (or all) of these lineages might have coincided with the viral induced RNA to DNA transition [35] and/or cellular lineages might have invented different antiviral mechanisms, such as specific type of virus-resistant outer membrane structures and/or envelopes, leading to their irreversible divergence [21,36,53].
If these various hypotheses cannot be easily tested, phylogenetic analyses have shown that viruses have at least shaped mitochondrial genomes, since the three major mitochondrial proteins, the RNA polymerase, the DNA polymerase and the DNA helicase, most likely originated from viral genes integrated into the genome of the alpha-proteobacterial symbiont [60]. This testifies that viruses have indeed shaped the genome of ribocell organisms in the distant past. More recently, it is now more and more recognized that retroviruses have played a major role in the evolution of modern cells [43]. For instance, it has been shown that viral infection has probably led to the emergence of the bacterial lineage phytoplasma [61]. In eukaryotes, repeated retroviral infections and their relics have introduced new proteins in the hosts proteomes and modified hosts regulatory networks with great impact on their evolution [62–64]. In particular, proteins of retroviral origin (syncitin) have played a crucial role in the evolution of mammals by providing a fusion mechanism for the formation of the placenta [65], and other viral-like proteins (recombinases) are probably at the origin of the immune system [66].
4.4 Toward a new synthesis in evolutionary biology
Darwinian evolution occurs via diversification and selection. At the time of Darwin, the source of variation was unknown. In the 20th century, the synthesis of Darwinism and genetics led to neo-Darwinism, which serves as a conceptual paradigm for the development of molecular biology. In this view, variations were mainly assimilated to mutations arising in the cellular genome. This explains why, for instance, some modern biologists have a problem to realize that extensive lateral gene transfer is fully compatible with Darwinism. We now realize that the source of variation is not limited to mutations and recombination, but includes lateral gene transfer (LGT) and even endosymbiosis. Although LGT is often considered as a mechanism to transfer cellular genes between cellular lineages, the vast majority of gene transfer corresponds in fact to the transfer in ribocells of genes synthesized in virocells [46]. In addition, viral integration promotes by itself mutations and recombination events. Viruses and derived elements, such as transposons and plasmids, are therefore the major source of genetic variation (the first pillar of Darwinism) through their ability in synthetic biology. But of course, viruses also put an enormous selection pressure on ribocells in their day-to-day life. Many studies have recently flourished showing that viruses control the number and type of cells in natural environments [67]. Survival to viruses (and vice versa, survival to ribocells counter-attack) is the key to the success of living lineages [43]. Ribocells have invented (or most likely recruited from viruses?) complex systems for their protection such as the modification/restriction and CRISPR systems in Archaea and Bacteria [68–70] and the RNA interference system in Eukarya [71]. So viruses are clearly not only the masters of variation but also the masters of selection (the second pillar of Darwinism). This led David Prangishvili and myself to propose recently that: “the billion years war between cells and viruses has been the major engine of life evolution” [20]. It is time now to integrate viruses in a new synthesis in biology that integrate in a sensitive way all living organisms in the grand scheme of Darwinian evolution [72].
5 Conclusion
Viruses have been discovered and first studied in the framework of the fight against infectious diseases. In the middle of the last century, they became suddenly model organisms for molecular biologists. Now, viruses are at the centre of discussions between evolutionists concerning their nature, their origin and their position in the tree of life [7,20,36,40,72,73]. Finally, they are also more and more involved in the design of new therapies (such as gene therapy) [74] or in the renewal of old ones (phage therapy) [75] and they have recently started to make their way in nanotechnology [76]. An aspect that has been neglected until now seems to be the more deliberate exploitation of their power as synthetic biologists. The concept of viruses as ribocells suggests focussing more on this aspect of virology. As suggested by the precedent of restriction enzymes, mining viral genomes for new biological functions, especially new tools to manipulate living organisms, is an avenue worth actively exploring in our new century.
Acknowledgment
I thank David Prangishvili for reading and correcting first versions of this manuscript.


