1 Introduction
Cellular metabolism is a complex process made of about a thousand chemical reactions catalysed by globular proteins, enzymes. As any other biological phenomenon, metabolism is the product of an evolutionary process. As the history and interrelationships of living organisms is based on comparative anatomy, the history of metabolism must be reconstructed by the comparative analysis of its structural complexity [1–4]. Biochemists recognized this necessity long ago, but have never used suitable comparative methods that would allow to test evolutionary hypotheses [2–8]. We propose the use of cladistic analysis [9,10] to infer the timing of emergences of a number of metabolic pathways, i.e. aliphatic amino acid catabolism and portions of the Krebs cycle, in order to shed light on the earliest among them.
1.1 Metabolic pathways and evolution
In 1945, Horowitz [11] postulated that the earliest biosynthetic pathways evolved in a backward direction if life began in a rich soup of organic molecules. If primitive cells were using a particular external nutrient, soon this organic molecule would be depleted in the environment. A selective advantage could be obtained by organisms able to synthesize this nutrient from an available precursor. Each biosynthetic step was selected according to successive depletions of precursors in the environment. The first enzyme to appear in the biosynthetic pathway was therefore the most distal in the pathway. Confluence of pathways was selected because it saved energy. This energy is used for other needs that will be more difficult to satisfy for competitor cells without confluence. This optimization of pathways is considered as a general basic rule of comparative biochemistry [1]. For these early anabolisms, common enzymes or common reactions shared by two (or more) synthetic pathways are distal, therefore evidence for common ancestry for these pathways. Pathways sharing these enzymes are closer to each other than to other pathways not using these enzymes.
In 1990, Cordón [2] proposed a symmetrical scenario of catabolic pathways. Early forms of life extracted energy from the degradation of substrates available in the environment into a product. Selective advantage was obtained for those able to produce a supplementary reaction of deeper degradation of this product, therefore obtaining more energy from the original substrate. Confluence is selected by obtaining the transformation of another substrate into an intermediate product already present in the protocell. The first reactions to appear in evolution of catabolism are proximal ones. The common distal elongation of two branched catabolic pathways is therefore a phenomenon whose final result will be evidence for common ancestry of these pathways. Two catabolic pathways sharing one or several distal portions of catabolism are supposed to be more closely related to each other than to other pathways. But there is a risk, which consist in the late branching of an ‘opportunist’ catabolic pathway when the early catabolism on which it branches is already complete. Common distal portion in this case is not an evidence for common ancestry, but just convergence obtained by recruitment, a phenomenon recognized for having played a role in biochemical evolution [5,12,13]. Homoplasy appears when there is character conflicts due to similarities obtained by evolutionary convergence or reversion. The risk of homoplasy in our data just depends on the relative timing between events of distal elongation of a pathway and the branching event of another one. An early branching event followed by distal elongation will provide good phylogenetic indicators. A late branching event (late in time and/or late in the pathway) will probably bring homoplasy. As in any other study of systematics where putative homologies are coded into a matrix, there are risks of homoplasy to carry on. We make the bet that this homoplasy will not swamp the phylogenetic signal.
According to Cordón [2], these rules are not rigid and can change from one type of metabolites to another: the order of development of a given pathway depends on the position and availability of the initial substrate and/or final product. The above timings in the genesis of reactions in anabolic pathways and catabolic pathways are valid because the final product and the initial substrate respectively are imposed from the outside to the cell, at least initially. Alternative scenarios can be obtained for transformations starting from products already integrated into the cellular metabolism. New biosynthetic pathways can develop in a forward direction by addition of new enzymes and reactions to preexisting pathways. For example, the urea cycle uses the biosynthesis of arginine [14]. Thus, Cordón [2] proposed a forward development for amino acid catabolism, fatty acids anabolism and glycogenesis, and a backward development for amino acid anabolism, fatty acids catabolism and glycolysis (confirmed in 1993 by Fothergill-Gilmore and Michels [15]).
So, it is clear that the evolutionary development of pathways has to do with the Darwinian concept of descent with modification. Present similarities in pathways detected through shared enzymes and enzymatic reactions can be interpreted as the result of pathway transformations through times. The comparison of pathways can therefore be followed by phylogenetic reconstruction, taking each catabolic pathway as a taxon, from its initial substrate to its entry into the Krebs cycle. For example, the taxon dASP1 is the catabolic pathway from aspartate to oxaloacetate, the enzymes and functions along this pathway will be the characters of this taxon (Fig. 1, characters 2, 13 and 31).

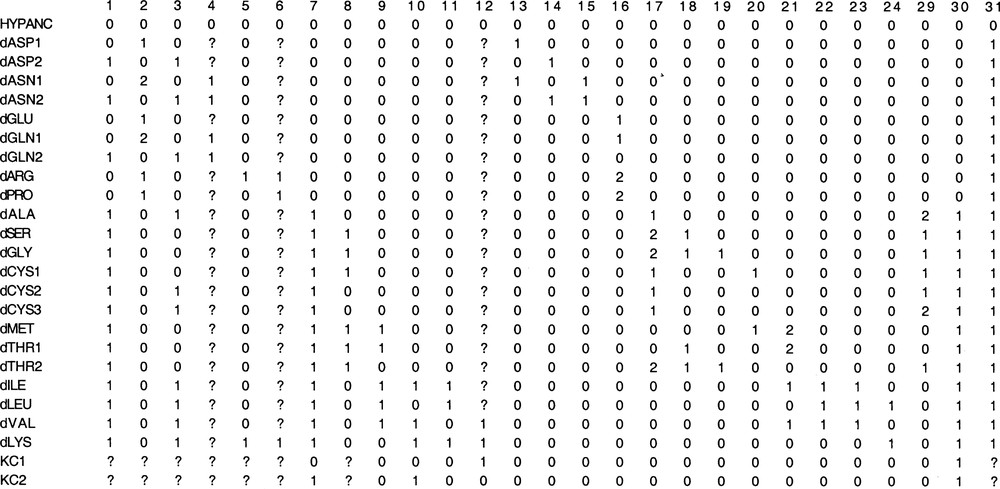
Matrix containing 25 taxons and 27 characters. Each taxon is a pathway of amino acid degradation (‘d'). Characters 25–28 and 32–58 have been removed because they only contain an autapomorphy. Characters are named above the matrix, with the corresponding number of international nomenclature [27], homology type, ordered or unordered.
1.2 The interest of amino acid catabolism
Amino acids are among the earliest abiotic sources of energy and molecules for protocells, without excluding other possible sources. Consequently, their catabolism is also a good candidate among the earliest biochemical reactions. First of all, it is classically admitted that catabolism must have preceded anabolism, even by a short time span. The main reason is that energy is required for anabolism, and the first source of energy is taken from catabolism because comes from the cleavage of some chemical bond of an abiotic molecule available in the environment. Biosynthetic pathways must have developed in parallel, but with a short delay. Why focusing on amino acids, and not on nucleic acids, fatty acids or monosaccharides? Reasons are many-fold. Free amino acids are found in abiotic environments like meteorites. Simple amino acids were obtained experimentally in an abiotic way, first by Miller in 1953 [16], who obtained glycine, alanine, glutamate and aspartate from simple molecules like ammoniac, hydrogen, methane and water. Serine can be synthesized abiotically in presence of formaldehyde. Moreover, among classical candidates for early energy providers (fatty acids, monosaccharides, amino acids), amino acids are the only chemical precursors whose structure is complex enough to contain all the atoms and reactive groups necessary for most of the reactions necessary for central metabolism. Other compounds like monosaccharides, polysaccharides, fatty acids have a poorer variety of atoms and groups and are rather monotonous. However, the strongest argument for early availability of amino acids to protocells might be the fact that any metabolism defined as a coordinated network of enzymatic activities performed by proteins need in its very early steps amino acid anabolism and catabolism for these proteins. Amino acid metabolism might therefore have preceded any other metabolism, even the most central metabolism like the Krebs cycle. But this question is debated.
1.3 The Krebs cycle
Whatever the metabolic specializations found in diverse living organisms (heterotrophy, photosynthetic autotrophy, chimiosynthetic autotrophy, various forms of respiration and fermentation), there is a universal core of about fifty metabolic pathways involving the anabolism and catabolism of amino acids, fatty acids, saccharides (the glycolysis and the glycogenesis, the pentose phosphate pathway) and the Krebs cycle. Because the Krebs cycle is the point of confluence of all other metabolic pathways, it is classically viewed as primitive. But this view is challenged by several lines of evidence. Molecules entering the Krebs cycle (oxo acids and acyl CoAs) are intermediate metabolites that are difficult to imagine available from primitive abiotic environments, but are most likely the products of a peripheral cellular metabolism. The origin of the Krebs cycle was thought to be secondary and composite by Schoffeniels [1,17], Gest [18,19], and Meléndez-Hevia et al. [7]. The later authors considered portions of the Krebs cycle as the evolutionary product of amino acid biosynthesis, instead of amino acid catabolism, because they excluded amino acid catabolism as a candidate right from the beginning. The main argument was that no pathway for amino acid and nitrogen-base degradation may have previously existed because “the selective value of a mechanism to eliminate organic material hardly built was not obvious at all”. The argument is circular because based on the assumption that amino acid anabolism pre-existed amino acid catabolism, because glycolysis was the first anaerobic source of energy (as in Gest [18]). We cannot follow this point of view: as catabolism must have preceded anabolism, it is more important to compare portions of the Krebs cycle to catabolism than to anabolism, when the question is to test for the earliest pathways between peripheral metabolism and portions of the Krebs cycle. Moreover, presence of glucose in abiotic conditions is less documented than presence of amino acids. Even if we do not exclude a priori the role of glucose as a first source of energy, this is not a sufficient reason to exclude the available amino acids. These considerations led us to incorporate two portions of the Krebs cycle as taxa in the matrix along with the catabolic pathways of each of the sixteen aliphatic amino acids in our data sets.
This work is the first cladistic analysis of the structure of metabolism. This is allowed by taking each pathway as a taxon and shared enzymes, shared enzymatic functions and shared cofactors as characters. Our aim is
- • to understand interrelationships of aliphatic amino acid catabolic pathways,
- • to discover in the most parsimonious tree the earliest metabolic pathway among them and portions of the Krebs cycle, and
- • to discover the first enzymatic functions to have occurred in protocells.
Comparing differences in metabolisms of extant living organism is of no help to reach this aim, because all the corresponding events are more differences in metabolism regulation than differences in structure of pathways [1], and, anyway, might have been posterior to the very early events we intend to infer. Comparing semantids of Zuckerkandl and Pauling [20], i.e. DNA sequences or protein sequences, is of no help because lead to severe problems of linear sequence homology and would infer mutational changes that also are posterior to the occurrence of these early metabolisms. At last, the present work infers nothing about information storage, replication and the RNA world.
2 Materials and methods
2.1 Sampling
Each aliphatic amino acid catabolic pathway is taken from the amino acid to its entry in the Krebs cycle and considered as a taxon. Some amino acid can be degraded through several possible ways (cysteine, aspartate, asparagine, glutamate, glutamine, threonine). In these cases each way is taken separately as additional taxons (for instance dCYS1, dCYS2, dCYS3 for the degradation (‘d’) of cysteine (CYS) following the pathways no. 1, no. 2, and no. 3 respectively). The Krebs cycle (KC) is considered in two portions, each beginning with an oxo acid which is a point of entrance into the cycle and also a point of output. These two oxo acids are also, among the metabolites of the Krebs cycle, the closest to amino acids, structurally speaking. The portion ‘KC1’ begins with oxaloacetate and ends with alpha-oxoglutarate; the portion ‘KC2’ begins with alpha-oxoglutarate and ends with oxaloacetate. Aromatic amino acids have not been considered at this stage of cellular evolution, because their complex metabolism needs too much oxygen [2] and is only possible once the metabolism of aliphatic amino acids is set.
2.2 Homology criteria
If shared enzymes or similar enzymes are evidence for common ancestry of metabolic pathways, similarities in the structure of active sites would be sufficient to formulate putative homologies. However, only a few active sites [21] are known in details, in comparison with the number of known enzymatic species [24]. We are therefore led to consider similarities in catalytic reactions and enzymatic mechanisms as a reflect of similarities in active sites. The higher the specificity, the more accurate this reflection is. In the same way, if we consider the generally accepted idea that enzymes evolved from low specificities to high specificities [5,12,22–25], putative common ancestry of pathways can be postulated not only on the basis of shared enzymes with high specificities, but also on the basis of very similar reactions, i.e. specificity in function but weak specificity for a substrate. To similarity in functions must correspond an underlying similarity in structure of active sites, a structural similarity that comes from common ancestry. Recognizing that two metabolic pathways share the same reaction with high specificity for substrate is using a strict criterion of primary homology [26], while recognizing a common family of reaction is relaxing this criterion, with a risk of homoplasy obtained by convergence or recruitment. There is no reason for considering that this risk is higher in the present case than the risk taken in primary homologies postulated in morpho-anatomical matrices usually constructed in systematics, or in aligned DNA sequences. The criteria of primary homology is fourfold: shared specific enzymatic activity (I), shared enzymatic function without shared specificity for substrate (IIa), shared coenzymes (IIb), and shared family of function (IIc).
2.2.1 Homology type I
Several pathways share the same enzyme with high specificity for its substrate. The enzyme itself is the hypothesis of primary homology. The absence is coded 0 and presence 1. For instance, the catabolisms of threonine and methionine both use the oxobutyrate dehydrogenase which transforms the oxobutyrate into propionyl coenzyme A. In both pathways the specificity of this enzyme for its substrate is the same. The character is called ‘oxobutyrate dehydrogenase’, it is coded 1 for taxa dTHR and dMET and 0 for dGLY, dLYS, etc. This criterion is used for characters 13–29, 32–58.
2.2.2 Homology type II
2.2.2.1 IIa: shared enzymatic functions
Several pathways utilize the same enzymatic functions, i.e. exhibit the same kind of chemical transformation, without considering the specificity of each enzyme for its substrate. The underlying hypothesis is that to similarity in enzymatic function must correspond similarity in structure of active sites, with the hypothesis that enzymes must have evolved from generalist active sites to specialized ones [12,22–25]. Relaxing in this way the criterion I allows transfering homology hypotheses to the past. When the substrate is present but the function not performed, the character state is coded 0. When the substrate is present and the function performed, the character state is coded 1. When the required substrate is not available, the character state is coded ‘?’. For example, catabolisms of alanine and aspartate perform transaminations using exactly the same enzymatic mechanisms, by two similar enzymes that differ in their specificities for their respective substrates: alanine aminotransferase and aspartate aminotransferase. The character state is coded 1 in dASP and dALA and other pathways where aminotransferases occur, 0 in dSER, dGLY, etc., and ‘?’ for portions of the Krebs cycle where transaminations are impossible. This criterion is used in characters 2–12.
2.2.2.2 IIb: shared cofactors
Shared cofactors reflect similarity in enzymatic mechanisms, which in this case do have the same functional meaning. If a common cofactor is used without similarity in enzymatic mechanisms, it is considered that the use of this cofactor has been gained independently, and each enzymatic mechanism is thus coded as homologies of type IIa. For example, this criterion does apply to the pyridoxal phosphate (PLP), which functional meaning is deamination. The character state is coded 0 when the deamination is not performed though possible, or performed but using another cofactor, 1 when a direct deamination or a transamination uses PLP, and ‘?’ for portions of the Krebs cycle where neither deaminations nor transaminations are possible.
This criterion is restricted in its application. First, when the use of a cofactor is too specific to an enzyme. Coding such a cofactor would lead to overweight the enzyme itself. For instance, the use of biotin is restricted to the propionyl carboxylase. In such a case the character ‘biotin’ won’t be taken into account. Second, when a cofactor is specific to an enzymatic function, there is a risk of overweighting a character of homology type IIa. For example, thiamin is specific to alpha-decarboxylations. Third, coding an ubiquitous cofactor brings risks of homoplasy. It is necessary in this case to consider the kind of reaction, that is, to come back to homology type IIa. For instance, NAD is used in a wide range of enzymatic functions: NAD-deaminations, NAD-aldehyde acid dehydrogenases, NAD-beta-oxydations, NAD-alpha-decarboxylations. Recoding each of these functions is useless: they are already coded as homologies IIa. It appears that this criterion is only useful for the PLP (character 1).
2.2.2.3 IIc: shared functional family
In its principle, this criterion is the same as in IIa, just relaxed. It is an extension of the above idea that enzymes must have evolved from generalists to specialists. The character state is coded 0 when the reaction is not performed though possible, 1 when the reaction is performed and ‘?’ when the reaction cannot be performed, considering chemical groups present in the metabolites of the pathway considered (‘?’ for deaminations in KC1 and KC2). This criterion actually concerns two main families of reactions: decarboxylations taken generally (character 30) and deaminations taken generally (character 31).
The original matrix contains 24 taxa (to which the hypothetical ancestor is added) and 58 characters. Characters 25–28 and 32–58 have been removed because autapomorphic, i.e. non informative for parsimony methods. Indeed, these characters exhibit a derived state in a single taxon. Consequently they cost a single step whatever the tree : they are not useful for discovering the most parsimonious tree among all possible trees. The final matrix contains 27 informative characters (Fig. 1).
2.3 Ordered characters
In some cases, several pathways share a couple of successive enzymes. Instead of coding these enzymes as separated type I homologies, their proximity have been incorporated into the matrix using ordered characters.
Some catabolic pathways use the pyruvate dehydrogenase for the transformation of pyruvate into acetyl CoA (dALA, dSER, dGLY, dCYS1, dCYS2, dCYS3; dTHR2). In some cases, (dALA, dCYS3), this step is preceded by the transformation of alanine into pyruvate, a reaction performed by alanine transaminase. Character 29: ‘alanine transaminase/pyruvate dehydrogenase’ is thus coded 0 when the pyruvate dehydrogenase is absent from the pathway considered, 1 when this enzyme is not preceded by alanine transaminase, 2 when pyruvate dehydrogenase is preceded by alanine transaminase. If this character is declared as ordered, the possible transformation 0 into 1 in the most parsimonious tree will have a cost of one step, while the possible transformation 0 into 2 will have a cost of two steps.
Alanine transaminase could have been considered as a single character of type I homology. But in our sample of pathways, this enzyme is always followed by the pyruvate dehydrogenase. The character ‘alanine transaminase’ must therefore be discarded to avoid overweighing it. This rule can be applied to all codings of successive enzymes: when successive enzymes have been coded using ordered characters, upstream enzyme must not be coded as an individual type I homology when it is always followed by the downstream enzyme. If it is not the case, the upstream enzyme can be left as an individual type I homology in the matrix. For example, in the couple serine deaminase-pyruvate dehydrogenase (ordered character 17), the upstream enzyme (serine deaminase) is not always followed by pyruvate dehydrogenase and is therefore left as a single character in the matrix (character 18). Other ordered characters are character 2 (‘amide deamination/deamination with NAD’), character 16 ‘aldehyde dehydrogenase/glutamate dehydrogenase’), and character 21 (‘oxobutyrate dehydrogenase/propionyl carboxylase’).
2.4 Tree reconstruction
2.4.1 Tree search
The most parsimonious tree was obtained through the ‘Branch-and-bound’ search of PAUP4 [28]. Trees are shown as phylograms, with branch lengths shown under ACCTRAN optimization.
2.4.2 Rooting
The tree was rooted using an all-zero hypothetical ancestor (HYPANC), according to the fact that, in the coding of character states, zero was given to absence of enzymes, to absence of performance of particular functions (even in presence of a putative suitable substrate), or to absence of utilization of a cofactor. We have to keep in mind that such a rooting option will automatically put the simplest pathways closer to the root. However, this does not make any assumption on the nature of the corresponding enzymatic reactions.
3 Results and discussion
3.1 Ordering the rise of enzymatic activities
The ‘Branch-and-bound’ search of PAUP4 [28] yielded 43 equiparsimonious trees of 46 steps (Consistency index (C.I.) = 0.7, Retention index (R.I.) = 0.85, strict consensus in Fig. 2). It is interesting to notice that character changes occurring on deep internal branches A, B, C, D, E are all uniquely derived characters (i.e. without homoplasy), except one (reversals for PLP transaminations). The temporal succession of these character changes provides the succession of occurrence of enzymatic catabolic activities in early protocells. First enzymatic catabolic activities must have been deaminations and aldehyde dehydrogenations (branch A), then decarboxylations (D, E). Within deaminations, deaminations of amids (A) must have preceded deaminations using either pyridoxal phosphate (C) or NAD (B). This has to be related to the fact that amide deamination does not use any cofactor. The earliest pathways to emerge are dASP1, dASN1, dGLU, dARG, dPRO, dGNL1. This group of pathways correspond to the first period of catabolic development as predicted by Cordón [2,3]. Then appear decarboxylations, the second period according to Cordón [2]. Beta-decarboxylations might have occurred early in this period (in KC1) because they do not use any cofactor, with two later convergences in dLYS and dVAL. Later on, transaminations are replaced by deaminations in pathways where the structure of the metabolite allows it (dSER, dGLY, dCYS1, dTHR2, dMET, dTHR1). The two blocks of the Krebs cycle appear to be the product of early amino acid catabolism (Figs. 2 and 3). KC1 emerges earlier in the tree than KC2 because KC2 needs alpha-decarboxylations (E) that were available later on. This does not mean that the complete pathway of KC1 was achieved before the complete KC2. KC1 is made of one beta-decarboxylation and other autapomorphic reactions which temporal organization is not deductible from the tree. KC2 shares some reactions with dLYS, dILE, dVAL, dLEU (beta-oxydations).
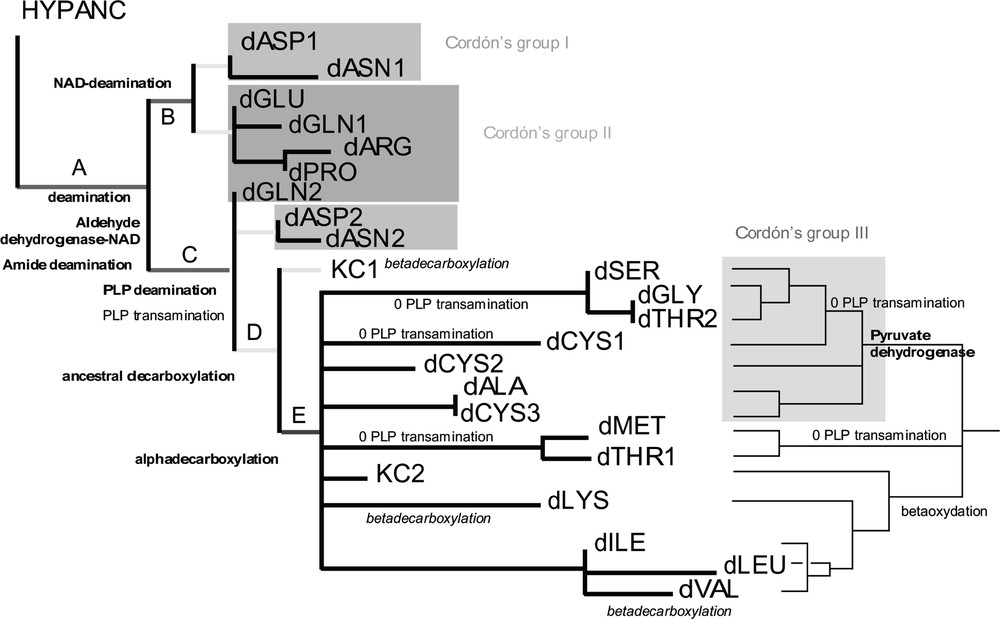
Left: a) strict consensus of 43 equiparsimonious trees obtained through a branch-and-bound search. Each tree is of 46 steps (C.I. = 0.7, R.I. = 0.85). Branch lengths are given under ACCTRAN. Only characters which changes occur at the deepest nodes A–D are shown. Character changes in bold are without homoplasy, in italics convergences, preceded by a zero reversals. Right: b) majority-rule consensus tree of the 43 trees. Only character changes useful for the text are shown. Other characters without homoplasy (not shown) are 14, 16, 17, 19, 21–23. Masquer
Left: a) strict consensus of 43 equiparsimonious trees obtained through a branch-and-bound search. Each tree is of 46 steps (C.I. = 0.7, R.I. = 0.85). Branch lengths are given under ACCTRAN. Only characters which changes occur at the deepest nodes ... Lire la suite

General view of amino acid catabolic pathways and their branching points in the Krebs cycle. Colours are successive time spans (or ‘epochs') as inferred from the cladogram: red first, then pink, deep blue, light blue, black. Reactions in black do not mean that all these reactions appeared at the same time. These are just reactions posterior to the blue epoch.
3.2 Nodes provide epochs of catabolic development
The cladogram provides a temporal sucession of enzymatic innovations as shown Fig. 3. In such a rooted tree (Fig. 2), nodes correspond to time spans in which enzymatic innovations occurred. These time spans or ‘epochs’ have been put in colour. The cladogram does not order events within an epoch but orders events between epochs. The first period (the red epoch and the pink epoch) is the period of deaminations and transaminations. Innovations occurring on red nodes preceded innovations occurring on pink nodes. In the red epoch, cysteine transforms into mercaptopyruvate, alanine into pyruvate, asparagine into aspartate, aspartate into oxaloacetate, glutamine into glutamate, glutamate into oxoglutarate (Fig. 3). Valine and isoleucine can be transformed into their respective oxo acids. During the next epoch (in pink), proline and arginine can be transformed into glutamate, and complete catabolisms of aspartate, glutamate, asparagine, glutamine, proline and arginine can be achieved. Then arrives the second period, in blue, when decarboxylations became possible (Fig. 3). The tree does not allow to distinguish any temporal succession between epoch in light blue (involving beta-decarboxylations) and epoch in deep blue (involving alpha-decarboxylations). Nevertheless, external arguments that will be discussed below allow to propose the deep blue epoch first, then the light blue epoch. In the deep blue epoch, branched oxo acids can be transformed into their respective acyl CoAs, pyruvate into acetyl CoA, oxoglutarate into succinyl CoA, oxobutyrate into propionyl CoA. In the light blue epoch, isocitrate is transformed into alpha-oxoglutarate, branched acyl CoAs into propionyl coenzyme A. The next phase (in black) are later steps among which the strict consensus tree cannot distinguish time spans further.
3.3 Events difficult to order
Beta-decarboxylations are simpler than alpha-decarboxylations because they do not require any cofactor and can easily be obtained spontaneously in acid environments. It is therefore convincing that they might have appeared first, for instance in the decarboxylation of oxaloacetate into pyruvate. However, the later reaction is not taken into account in our matrix because it belongs neither to amino acid catabolism nor to the Krebs cycle sensu stricto. Among the pathways considered here, all the metabolites produced at the end of the first period of deaminations are alpha oxo acids. Only alpha-decarboxylations were thus possible in the next step (deep blue), and beta-decarboxylations just after as soon as possible. This is the reason why alpha-decarboxylations have been coloured in deep blue in Fig. 3, distinguished from beta-decarboxylations in light blue.
Although KC1 in the tree is branched earlier than KC2, this does not mean that the complete achievement of KC1 occurred before the complete achievement of KC2. Precedence of KC1 over KC2 cannot be definitively established here without taking into account amino acid anabolic pathways. Indeed, if we had taken anabolism into account, it is not excluded that some anabolic pathways would have been branched among certain catabolic pathways. Moreover, in a different interpretative context taking anaerobic glycolysis a the first source of energy, Gest [18,19], Meléndez-Hevia et al. [7] described the process by which KC1 would have arisen through the biosynthesis of glutamate, from pyruvate to glutamate. Since this pathway was not coded as such, we won’t provide firm conclusions at the moment about the precedence of KC1 over KC2.
3.4 Corroborating Cordon’s scenario
Cordón [2] distinguished four groups of amino acids based on the structure of catabolic and anabolic pathways [3]. The first phase of evolution according to Cordón is the period of deaminations and transaminations (red and pink) leading to the development and the complete achievement of the pathways of amino acids of groups ‘I’ (dASP1, dASN1, dASP2, dASN2 ,) and ‘II’ (dGLU, dGLN1, dARG, dPRO, dGLN2). These two groups are paraphyletic but do appear as the most basal ones. In the second phase, decarboxylations are added (blue), during which would have developped until their present structure catabolic pathways of amino acids of Cordón’s group ‘III’ (dSER, dTHR2, dGLY, dCYS1, dCYS2, dCYS3, dALA). In the strict consensus of our 43 equiparsimonious tres (Fig. 2, left), this group does not appear because of collapsed nodes. However, in the majority-rule consensus tree (Fig. 2, right), Cordón’s group III is monophyletic and supported by ordered characters 17 and 19 involving the use of pyruvate dehydrogenase (without homoplasy). Last, Cordón considered a third phase during which occurred complementary complex reactions and the complete closing of the Krebs cycle. This phase corresponds in the majority-rule consensus tree to two clades, the one grouping dMET and dTHR1, and the one based on shared beta-oxydations grouping KC2, dLYS, dLEU, dILE et dVAL (with a beta-oxydation reversal in dLEU).
It is also interesting to confirm Cordón’s views on the direction of catabolic pathways’ evolution. The first reactions of amino acid degradation in the pathway (deaminations, transaminations) are the first in the course of evolution: deaminations and transaminations occur at the deepest nodes of our tree. One must keep in mind that none of Cordón’s ideas have been taken into account in the coding of character states. One could object that the obtained result depends of our rooting option. Indeed, the simplest pathways are basal because of the all-zero hypothetical ancestor. But this makes no assumption on the nature of the simplest pathways. The fact that the simplest pathways contain only deaminations (and not, for instance, decarboxylations) is not a matter of arbitrary rooting but just a fact written in the structure of these catabolic pathways.
3.5 Recruitment of enzymes and opportunist late branching of pathways
Pathways can evolve by recruitment of enzymes or portions of pathways, so that depicting interrelationships of pathways under the form of a tree might be inappropriate. Such processes should typically provoke homoplasy in trees by opportunist late branching of pathways, at least detectable by the decrease of the C.I. and the R.I. However, values obtained for C.I. and R.I. are rather high, not compatible with the idea of complete reticulate mode of evolution. In the same way, one could have been discouraged of using trees because of the possibility of gaining the same enzymatic functions from different structures, or because similar enzymatic mechanisms could have arisen a number of times. Nevertheless, this risk is the burden of any comparative approach, and our homoplasy measurments indicate that these homoplastic events have not been dominant.
3.6 Methodological interest
By proposing a new kind of taxon, the catabolic pathway, and using the enzymes, enzymatic functions, cofactors and families of enzymatic functions as characters, it is possible to propose the first phylogeny of some (but obviously not all) metabolic pathways. This phylogeny independently confirms the views of Cordón [2] who did not use cladistic analysis. This work sets a general methodological framework within which other metabolic pathways now can be analysed.
Acknowledgements
We thank Marc Silberstein and Patrick Tort for their help. The Institut Charles Darwin International, the Muséum National d’Histoire Naturelle, and La Maison des Sciences de l’Homme are acknowledged for fundings to C.C.
Version abrégée
Une des toutes premières sources d’énergie des premiers systèmes vivants a pu résider dans des acides aminés aliphatiques libres. De ce postulat découle l’idée que les voies de dégradation des acides aminés aliphatiques sont de bons candidats pour figurer parmi les premières voies métaboliques du vivant et qu’elles seraient pour une bonne part à l’origine du cycle de Krebs. Lorsqu’il s’est agi de discuter cela et d’inférer les premiers tronçons d’un métabolisme primitif, les biochimistes ont conçu des scénarios avec de bon arguments mais sans méthode d’inférence précise. Pour contribuer à cet effort, une analyse cladistique des voies de dégradation des seize acides aminés aliphatiques et de deux portions du cycle de Krebs a été menée. L’objectif était triple : premièrement, identifier la (les) voie(s) de dégradation d’émergence la (les) plus précoce(s), deuxièmement, trouver l’ordre temporel d’apparition des grandes fonctions enzymatiques dégradatives, troisièmement, vérifier si le cycle de Krebs (ou certaines de ses portions) est antérieur aux voies de dégradation des acides aminés ou bien s’il n’est qu’un produit de celles-ci. L’investigation repose sur deux postulats : 1. les voies métaboliques ont une histoire interprétable en termes de transmission dans le temps avec modification, et 2. lorsque les voies de dégradation de deux acides aminés emploient le même enzyme ou le même groupe d’enzymes successifs, nous parions qu’à cette portion commune de voie métabolique correspond une ascendance commune. Ce critère d’homologie primaire est également étendu aux fonctions enzymatiques, cofacteurs et familles de fonctions. En d’autres termes, en utilisant les voies de dégradation de chaque acide aminé aliphatique comme taxon, les enzymes et réactions enzymatiques comme nouveaux caractères, et un ancêtre hypothétique, nous pouvons formuler des homologies putatives (ou homologies primaires) par une matrice de caractères et inférer pour la première fois une phylogénie des voies du catabolisme. Le branchement des voies postérieur à leur mise en place, par recrutement, constitue un risque d’homoplasie qui est discuté. Le cladogramme résultant montre une émergence précoce des voies dégradatives de l’aspartate et de l’arsparagine, des glutamate, glutamine, proline, et arginine. Dans le consensus majoritaire, toutes les voies utilisant la pyruvate déshydrogénase (groupe III de Cordón, 1990) apparaîssent apparentées entre elles. Les premières réactions enzymatiques ont dû être des désaminations, les transaminations, puis des désaminations utilisant le pyridoxal phosphate, puis ensuite vient la phase de l’histoire catabolique où les décarboxylations sont possibles. L’ordre d’apparition des différentes fonctions enzymatiques permet de discriminer quatre périodes de temps, ou fenêtres temporelles, indiquées sur le schéma général des voies cataboliques des acides aminés. Cette étude pose les jalons méthodologiques d’une biochimie comparative moderne, c’est-à-dire connectée avec les concepts, méthodes et outils de la systématique.


