1 Introduction
Plasma high-density lipoprotein (HDL) plays a key role in maintaining cholesterol homeostasis. SR–BI is a member of the CD36 superfamily of genes. This high-affinity HDL receptor mediates the selective uptake of HDL cholesterol [1]. SR–BI is most highly expressed at all major sites of selective cholesterol uptake, such as the adrenal glands, the ovaries and the testis, all of which produce steroid hormones, and also in the liver, which is involved in the transfer of excess cholesterol from the peripheral tissues by a process termed ‘reverse cholesterol transport’. This process helps to decrease the plasma cholesterol concentration and thus counteracts atherosclerosis [2]. Furthermore, SR–BI is expressed in atherosclerotic plaques and in human retinal pigmentary epithelium (RPE) cells [3,4], suggesting that it is involved in the uptake of native or oxidized photoreceptor outer segment (POS) lipids from the RPE and in the phagocytosis of POS by RPE cells. The metabolism of photoreceptor lipids depends, at least partially, on RPE cells, which display a scavenger receptor activity similar to that of macrophages and arterial endothelial cells [5]. This receptor is also expressed in monocytes and macrophages, where it may act as a scavenger by binding to oxidatively modified low-density lipoproteins, senescent or apoptotic cells [6], and anionic phospholipids [7,8]. It has long been thought that RPE cells are major targets of pathogenic processes, leading to the formation of drusen and age-related macular degenerations (AMDs). AMDs are the leading cause of visual impairment in the elderly and the most common causes of blindness in the developed world. There is a strong inverse correlation between HDL cholesterol levels and the risk of coronary disease and the risk of AMD [9]. Cholesterol accumulates in Bruch's membrane in humans with age [10], and in a similar way in the intima of atherosclerotic arteries, where lipoproteins are the source of extracellular cholesterol. Given these observations, we decided to study the ocular expression pattern of the SR–BI gene, with particular emphasis on retinal expression. The possible implication of SR–BI in retinal degenerations also led us to compare its expression in the RCS rdy– rat eye and in the normal RCS rdy+ eye. The RCS rdy– strain of rats presents an autosomal recessive retinal dystrophy characterized by the abrogation of POS phagocytosis by abnormal RPE cells. The abrogation of the ingestion phase of POS phagocytosis is caused by the homozygous deletion of the c-mer gene. This triggers a secondary photoreceptor degeneration that characterizes the RCS rdy– rats. These rats constitute a model of Retinopathy Pigmentary caused by a primary defect of the RPE and may provide important clues concerning the pathophysiology of some forms of AMD. CD36 has been demonstrated to be down regulated in the RPE of RCS rdy– rats as compared to that of RCS rdy+ rats [11]. Furthermore, recently CD36 ligation has been demonstrated to be necessary and sufficient for activating the ingestion phase of POS by RPE cells [12]. We then hypothesized that SR–BI, a CD36 homologue could have also a role yet to be determined in the phagocytosis of POS by RPE cells and could be differentially modulated in the RPE of strains of RCS rats as compared to control.
Cholesterol is a key molecular component of all cell membranes and is also the primary substrate in steroidogenesis. Steroids are synthesized de novo from cholesterol by specific enzymes (e.g., cholesterol side-chain cleavage cytochrome p450, P450scc, 3β-hydroxysteroid dehydrogenase, 3β-HSD) located in the mitochondria. Cholesterol is transported into mitochondria by the steroidogenic acute regulatory protein (StAR). StAR mediates the rapid transfer of cholesterol from the outer to the inner mitochondrial membrane where it can be cleaved into pregnenolone, the first steroid hormone formed [13]. The transfer of cholesterol is the rate-limiting step of steroidogenesis. Neurosteroidogenesis occurs in brain and retina [14–18]. Another aim of this study was to determine the localization of the StAR protein in rat ocular tissues, with special emphasis in the retina to determine the cellular sites of the eye where SR–BI and StAR may be co-expressed.
2 Materials and methods
2.1 Animals
The RCS rdy+, p+ rat strain was used as the reference strain. The RCS rdy–, p+ rat strain was also used. Animals were reared at 20 °C in a 12 h light/12 h dark cycle.
2.2 Tissue sampling
Adult rats (2 months old) were killed by CO2 asphyxiation. Eyes were rapidly removed and fixed for at least 36 h in 4% PFA at 4 °C. They were then embedded in paraffin and cut into 5-μm sections.
2.3 RNA and protein extractions
Rat tissues were rapidly removed and immediately frozen at −80 °C. Before freezing, the eyes were microdissected such that the major part of the retina was detached from the eye, leaving the RPE attached to the choroid. The RPE was then gently scraped from the choroid with a razor blade.
Total RNA and proteins were extracted from tissues with the Qiagen RNA Extraction Kit, and TRIzol reagent (Life Technologies) respectively, according to the manufacturer's instructions.
2.4 Semi-quantitative reverse transcription-polymerase chain reaction (RT–PCR)
Total RNA (2 μg) was reverse transcribed using the First-Strand cDNA synthesis kit (Amersham) according to the manufacturer's recommendations. SR–BI was PCR-amplified using the following primers (0.15 μM): gagcgtggaccctatgtcta (forward) and ggtgctgacattctgaatgcc (reverse). Cyclophilin (internal control) was amplified in the same reaction with the following primers (0.30 μM): tggtcaaccccaccgtgttcttcg (forward) and tccagcatttgccatggacaaga (reverse). The expected sizes of the PCR products are: 311 bp for cyclophilin and 738 bp for SR–BI. RT–PCR was carried out for 25 cycles.
We measured the optical densities of the PCR bands using NIH Image (http://rsb.info.nih.gov/nih-image/). The relative levels of mRNA were then calculated as follows: optical density of SR–BI band/optical density of cyclophilin band.
2.5 Western blot analysis
The extracted total proteins (80 μg), were separated onto a 15% SDS-PAGE gel and were transferred onto a nitrocellulose membrane (Protran BA 83, Schleicher & Schuell). Membrane was incubated as control with a β actin polyclonal antibody (1:2000 dilution; Santa Cruz Biotechnologies) for 2 h at room temperature (internal control) and incubated with horseradish peroxidase-conjugated anti-goat IgG ( dilution, Sigma) at room temperature for 1 h. Secondly, the blots were incubated, at 4 °C overnight, with the RED-1 antibody (1:1000 dilution) which recognizes SR–BI and SR–BII proteins (Novus Biologicals NB 400-102) and has already been thoroughly validated for its specificity [19]. The blots were incubated with horseradish peroxidase-conjugated anti-rabbit IgG ( dilution: Amersham) at room temperature for 2 h. ECL Western blot analysis system (Amersham Pharmacia Biotechnologies) was used to detect bound antibodies.
2.6 In situ hybridization
The 738-bp SR–BI cDNA extracted from rat liver was subcloned with the pGEM-T-easy vector system II kit (Promega). Sense and anti-sense riboprobes were synthesized by in vitro transcription with T7 and SP6 RNA polymerases (Roche Molecular Biochemicals), respectively, using DIG-labeled uridine triphosphate (DIG RNA Labeling Kit, Promega).
The DIG-labeled cRNA probes were hybridized to the pre-treated tissue sections overnight at 65 °C in a hybridization buffer and the sections were then incubated with an alkaline phosphatase-conjugated anti-digoxygenin antibody (AP-Fab fragments Roche Molecular Biochemicals) at a dilution of 1:2000 in PBS-Triton (with 1% goat serum) at 4 °C overnight. The sections were stained with an NBT (100 mg ml−1)/BCIP (50 mg ml−1) solution (NBT-BCIP kit, Roche Molecular Biochemicals) in NTMT coloring buffer and coverslipped (with an aqueous mount medium, Aquatex Merck). SR–BI mRNA appeared as a dark blue precipitate within cells.
2.7 Immunohistochemistry
The cellular localizations of the SR-B proteins in the eyes of adult rats were analyzed by immunohistochemistry with the DAKO ChemMate peroxidase/DAB detection kit. The RED-1 antibody 1:2000 (Novus Biologicals NB 400-102) recognizes SR–BI and SR–BII proteins [19] and the rabbit-anti-mouse StAR antibody (dilution 1:300; provided by Dale Hales, University of Illinois, Chicago) is specific for the recognition of the StAR proteins and has been also thoroughly validated [20]. Tissue sections pre-treated and depigmented were incubated with antibodies for 1 h, with the peroxidase staining kit for 30 min and counterstained with 3% Methyl Green solution (Sigma).
3 Results
3.1 SR–BI transcripts are present in rat eye and brain tissues
We used semi-quantitative RT–PCR to examine the relative levels of SR–BI mRNA in adult rat tissues, after normalization to cyclophilin. RT–PCR detected a 738-bp product in both in eye tissues (RPE, neuroretina), cerebral tissues (cerebellum, forebrain) and all three-control tissues (ovary, testis, liver) from RCS rdy+ rats (Fig. 1A). An intense band was detected in the brain, neuroretina and RPE. SR–BI mRNA levels were highest in the ovary (Fig. 1B). Interestingly, the test tissues (retina, brain) appeared to contain the same amount of SR–BI mRNA as the ovary. This experiment was carried out in triplicate and no significant variation was observed between each experiment. We obtained similar results with RCS rdy– tissues (data not shown).

RT–PCR semi-quantitative analysis of SR–BI mRNA transcripts in rat tissues. Total RNAs were isolated from rat ovaries, liver and testis (positives controls); from heart, cerebellum, forebrain, RPE and neural retina. (A) RT–PCR was carried out with primers specific for rat SR–BI mRNA after cDNA concentration was normalized to cyclophylin gene expression. PCR products of the expected length (738 pb) were amplified. (B) Densitometric analysis of intensities of PCR bands. The relative levels were calculated as the ratio of the SR–BI intensity band on cyclophylin intensity band. Peak intensities of the PCR bands occurred in brain, RPE and neuroretina (n=3). Error bar, SE.
3.2 SR–BI proteins are synthesized in the rat retina
We subjected proteins extracted from the neuroretina and RPE of adult RCS rdy+ rats to western blot analysis using the RED-1 antibody. This antibody recognized an 82-kDa protein in both retinal tissues (Fig. 2). This result is in good agreement with a previous western blot analysis that detected the SR–BI receptor in a protein homogenate extracted from primary human RPE cultures [3]. Thus, SR–BI receptors are expressed in the rat neuroretina and RPE. We also used western blotting to analyze the production of SR–BI in the neuroretina and the RPE of RCS rdy– rats (data not shown). We did not observe any variations in the amount or the size of the proteins detected in the tissues of the two types of rat strains investigated.

Western blot analysis for SR–BI proteins. Western blot analyses of rat neural retina and RPE membrane were performed with the affinity antibodies RED-1 and β actin polyclonal antibody (internal control). Crude membrane (80 μg) was applied to 10% SDS-polyacrylamide gel. Specific bands for SR–BI (82 kDa) and β actin (43 kDa) were detected in neural retina and in RPE protein extracts.
3.3 SR–BI transcripts are present in many retinal cell layers and in other ocular tissues
We used in situ hybridization (ISH) analysis to determine which retinal cells and which neuronal and non-neuronal ocular tissues contained mRNA encoding SR–BI. SR–BI mRNA appeared as a dark blue precipitate within cells (Fig. 3). SR–BI mRNA was detected in several cell layers of the neural retina and in choroidal and scleral cells (Fig. 3B). It was detected in the inner nuclear layer (INL), outer nuclear layer (ONL), photoreceptor inner segments, RPE cells and endothelial cells of both the retina and choriocapillaris (Figs. 3 B–E). The examination of higher magnification images of the inner retina (Fig. 3C) revealed the presence of large amounts of SR–BI mRNA in retinal ganglion cells, the nerve fiber layer, amacrine cells, and cells located in the INL. SR–BI mRNAs were also detected in the oligodendrocytes of the optic nerve (data not shown).

Distribution of SR–BI specific mRNA transcript in adult rat eye detected by in situ hybridization. The intense purple stain indicates a positive reaction. In situ hybridization, using specific riboprobes prepared from non-coding regions of SR–BI cDNA, demonstrated the existence of SR–BI mRNA in all nuclear layers of the retina (B–E). Several layers of the retina are indicated (GCL, ganglion cell layer; A, axon; INL, inner nuclear layer; ONL, outer nuclear layer; IS, inner segment, OS, outer segment; RPE, retinal pigment epithelium). Non-retinal tissues showed positive reaction in (E) choriocapillary endothelial cells (Ch). (F), ciliary body. Strong signals for SR–BI mRNA were observed in pigmentary ciliary epithelium (PCE) and non-pigmentary ciliary epithelium (NPCE). (G), iridocorneal angle. There was a stronger staining in the iris stroma (IrS), and in the posterior iris pigmentary epithelium (PIPE) than in the trabecular meshwork (TM). (H), Cornea. Strong signals for SR–BI mRNA were observed in corneal epithelial (CEp), corneal stromal (S), and corneal endothelial cells (CEn). (I), lens. The lens epithelium (LE), and lens equator showed a positive reaction. Capsule (CP). (A) rat retina probes with sense (negative control) digexygenin labelled SR–BI riboprobe. No specific signal is detected with the sense probe. V, Vs: vessels. Sc: Sclera. Bar, 50 μm.
Non-neuronal ocular tissues also contained mRNA encoding SR–BI. The pigmented and non-pigmented ciliary body epithelial layers were both positive for SR–BI mRNAs (Fig. 3F). Most cells of the iris stroma, iris epithelium and some cells of the trabecular meshwork contained SR–BI mRNAs (Fig. 3G). In the cornea, the columnar cells of the stratified epithelium, the stromal keratocytes and the corneal endothelium were positive for SR–BI mRNAs (Fig. 3H). In the lens, both the epithelial cells of the intermediate zone and lens equator were positive for SR–BI mRNAs (Fig. 3I). The lens anterior epithelium and the nascent nucleated lens fibers were strongly labeled whereas the non nucleated lens fibers displayed positive but moderately intense SR–BI signals. We did not find any differences between the SR–BI mRNA patterns in normal RCS rdy+ and RCS rdy– eyes (data not shown).
3.4 Immunolocalization of SR–BI in rat ocular tissues
The in situ hybridization data showed that SR–BI mRNA is present in all ocular tissue cell types. We then wanted to know whether the cells also contained the SR–BI protein. SR–BI proteins were detected in ganglion cell layer and in thin areas surrounding the somata of the cells in the inner nuclear layer (Fig. 4B). These thin areas are probably membranes of the bipolar neurons and possibly of the Müller cells. Examination of the inner retina at a higher magnification (Fig. 4C) revealed intense labeling of the retinal ganglion cells and of their axons. Oligodendrocytes and optic nerve fibers were also labeled strongly (data not shown). SR–BI was also detected in both the inner and outer plexiform layer (IPL and OPL respectively) (Figs. 4C and 4D), in the inner segments of photoreceptors and in the RPE layer (Fig. 4E). In non-retinal tissues, SR–BI was detected in scleral cells (Fig. 4E). The choroid was very strongly labeled by the RED-1 antibody. The endothelial cells of the choroid and those of the choriocapillaris were intensely labeled for SR–BI (Fig. 4E). High levels of protein were detected in corneal epithelial cells, stromal keratocytes and corneal endothelial cells (Fig. 4H). Most iris cells, including iris pigmented cells, melanocytes and capillary endothelial cells displayed intense SR–BI immunoreactivity (Fig. 4G). High levels of SR–BI were observed in the epithelial cells of the ciliary body (Fig. 4F), in the equatorial cells and in the anterior epithelial cells of the lens (Fig. 4I). Strong SR–BI immunoreactivity signals were observed in all lens epithelial fibers. Finally, the ocular skeletal muscle cells also contained SR–BI (data not shown). We did not find any differences between the distribution of the SR–BI protein in normal and RCS rdy– eyes (data not shown).
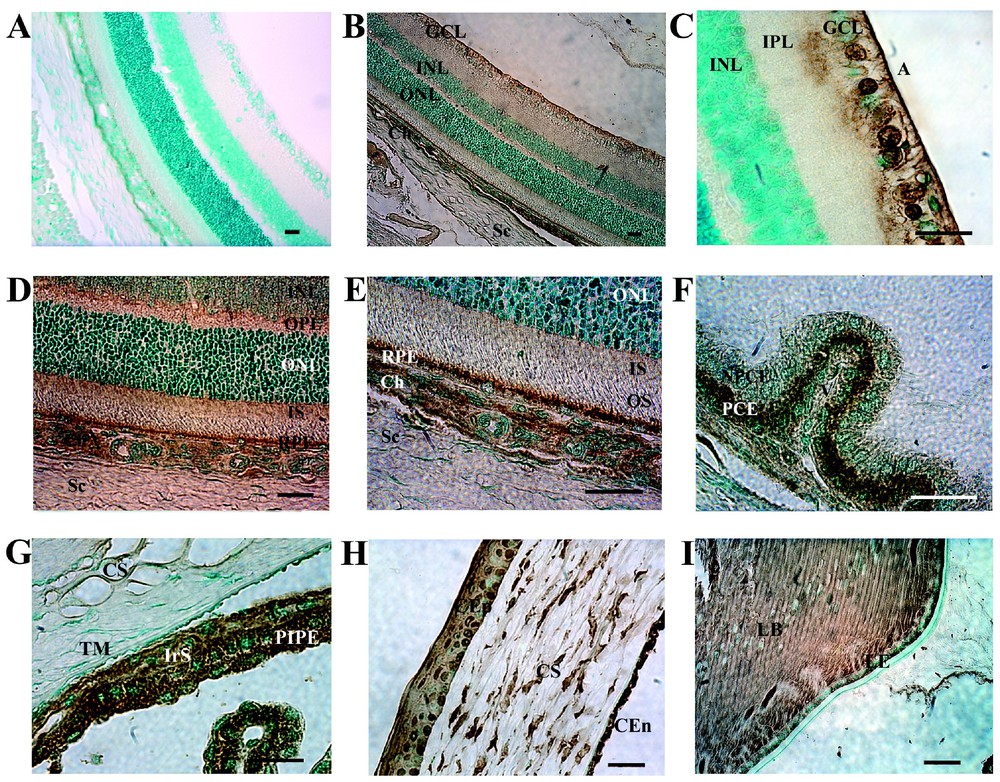
Immunolocalization of SR–BI in adult rat eye. Intense immunoreactivity was observed in several layers of the retina (B–E) in the ganglion cell layer (GCL), nerve fiber layer, inner and outer plexiform layers, photoreceptor inner segment (IS) and retinal pigment epithelium (RPE) cells. Faint immunoreactivity was detected around soma of cells in the inner nuclear layer (INL) and outer nuclear layer (ONL). In non-retinal tissues, SR–BI protein was strongly observed in (E) choriocapillary endothelial cells (Ch). (F) Ciliary body. SR–BI protein was strongly detected preferentially in pigmentary ciliary epithelium (PCE) than in non-pigmentary ciliary epithelium (NPCE). (G) Iridocorneal angle. An intense specific immunolabelling of the iris stroma (IrS) and posterior iris pigmentary epithelium (PIPE) was observed. There is a faint immunoreactivity in the trabecular meshwork (TM). (H) High magnification of the cornea showing intense immunolabelling of corneal epithelial (CEp), and some labelling of the corneal stromal (S), and corneal endothelial cells (CEn). (I) Lens. Note the immunolabelling of cells along the equator of the lens and in the lens epithelium (LE). (A) Immunonegative control for SR–BI. A, axon; OS, outer segment. Bar, 50 μm.
3.5 The steroidogenic acute regulatory (StAR) protein, a key enzyme in steroidogenesis, is colocalized with SR–BI in the eye
We compared the distribution of the StAR protein in ocular tissues with that of SR–BI. At a low magnification (Fig. 5B), StAR and SR–BI were detected in the same cells: in the ganglion cell layer (Fig. 5C) and in the thin layer surrounding all cell somata in the inner nuclear layer. This suggests that both bipolar neurons and Müller cells may produce StAR. Like SR–BI, moderate levels of StAR were detected in the IPL and high levels were detected in the OPL and in the photoreceptor inner segments (Figs. 5D and 5E). High levels of StAR were present in the RPE cell layer and in the choroidal layer (Fig. 5E). Examination of the inner retina at a higher magnification (Fig. 5C) revealed intense labeling of retinal ganglion cells, oligodendrocytes and astrocytes. StAR and SR–BI appear to display a similar distribution very likely to be identical in the extra-retinal tissues of the eye (Figs. 5 F–I). These results suggest strongly a colocalization of StAR and SR–BI in the same ocular cells but additional double immunostaining experiments are needed in order to confirm this hypothesis.
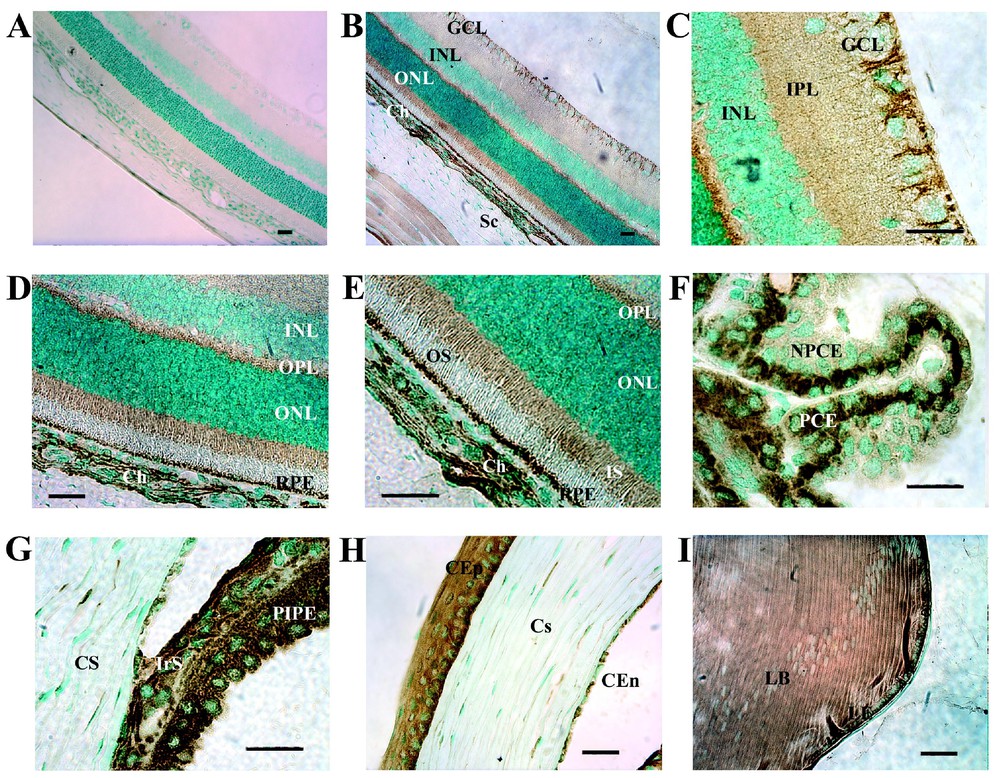
Immunolocalization of StAR in adult rat eye. Intense immunoreactivity was observed in several layers of the retina (B–E) in the ganglion cell layer (GCL), nerve fiber layer, inner and outer plexiform layers, photoreceptor inner segment (IS) and retinal pigment epithelium (RPE) cells. Immunoreactivity was detected around soma of cells in the inner nuclear layer (INL) and outer nuclear layer (ONL). In non-retinal tissues, StAR protein was strongly observed in (E) choriocapillary endothelial cells (Ch). (F), ciliary body. StAR protein was more strongly detected in pigmentary ciliary epithelium (PCE) than non-pigmentary ciliary epithelium (NPCE). (G) Iridocorneal angle. An intense immunolabelling specific of the iris stroma (IrS) and posterior iris pigmentary epithelium (PIPE) was observed. There is a faint immunoreactivity in the trabecular meshwork (TM). (H) High magnification of the cornea showing intense immunolabelling of corneal epithelial (CEp), and some labeling of the corneal stromal (S), and corneal endothelial cells (CEn). (I) Lens. Note the immunolabelling of cells along the equator of the lens, in the lens epithelium (LE). (A) Immunonegative control for StAR. OS, outer segment. Bar, 50 μm.
4 Discussion
Our results demonstrate for the first time that SR–BI mRNAs and proteins are abundant in ocular localizations characterized by a high level of membrane production and renewal. Most of the cholesterol is localized in the IPL, but is largely incorporated into the newly synthesized discs at the base of POS [21]. SR–BI mRNA and proteins are produced in the retina, particularly in the photoreceptor inner segments (PIS), one of the subcellular photoreceptor cell areas with the highest density of mitochondria. The widespread presence of SR–BI in ocular tissues might reflect the need for cells that have to renew their membranes continually and thus to receive a constant supply of cholesterol and other lipids. Indeed, the cornea and the lens are also major sites of continuous membrane biosynthesis, requiring a constant supply of cholesterol [22].
It is noteworthy that large amounts of SR–BI mRNAs and proteins are present both in the outer blood-ocular barrier, primarily constituted by RPE cells, and in the inner blood–ocular barrier, constituted by inner endothelial cells surrounded mainly by astrocytes. RPE cells have two fundamental properties: the supply of retinoids and vitamin E to the photoreceptor cells, and the phagocytosis of the ‘older’ POS disks.
The visual pigments of animals are composed of a retinoid chromophore, which is a carotenoid derivative, bound to a protein moiety (opsin) embedded in the photoreceptor membranes. A class B scavenger receptor, ninaD, mediates the cellular uptake of carotenoids in Drosophila [23] and shares significant sequence identity with the mammalian class B scavenger receptors, SR–BI (24% sequence identity) and CD36 (22% sequence identity). The Drosophila ninaD mutant is blind due to a nonsense mutation in the ninaD gene that leads to a defect in carotenoid utilization for the synthesis of the visual chromophore. There is growing evidence that class B scavenger receptors, in particular SR–BI, are involved in lipid metabolism, especially cholesterol homeostasis in mammals as exemplified by knock-out and transgenic mice [24,25]. We found that SR–BI, similarly to its ninaD ortholog, is expressed in the photoreceptors. However, no defects related to carotenoid metabolism have yet been reported in SR–BI- or CD36-deficient mice. Vitamin E is also essential for normal neuronal and retinal functions as chronic vitamin E deficiencies result in characteristic and severe neurological diseases such as Friedreich's ataxia and pigmentary retinopathies [26]. Vitamin E (alpha-tocopherol, αTocH) is a lipid-soluble antioxidant [27], it is transported in association with lipoproteins in the peripheral circulation. SR–BI is expressed in porcine brain capillary endothelial cells constituting the blood-brain barrier and is involved in the selective uptake of HDL-associated vitamin E [28]. SR–BI mediates the selective uptake of HDL-associated αTocH in a similar manner to that described for HDL-associated cholesteryl esters. The metabolism of αTocH is abnormal in SR–BI-deficient mice, resulting in an increased plasma concentration of this fat-soluble vitamin [29]. The defective tissue uptake of αTocH by SR–BI-deficient mice (ovary, testis, lung and brain) may contribute to the reproductive and cardiovascular diseases exhibited by these animals [30]. These data highlight the necessity to monitor more carefully the visual functions and the histopathology of SR–BI-deficient mice at all stages of their life. Our observation that retinal and choroidal endothelial cells contain high levels of SR–BI mRNAs and proteins, especially the choriocapillaris endothelial cells and the other endothelial and smooth muscle cells of ocular vessels, including those of the ciliary body and iris, is in accordance with the data collected in the brain vessels [31].
RPE cells are directly involved in the phagocytosis of shed POS discs. SR–BI is expressed by RPE cells. Although SR–BI is involved in the clearance of apoptotic cells [32–34] and in the clearance of apoptotic astrocytes cells in vivo [35,36], our study did not reveal any differences between the expression of SR–BI in the normal RCS rdy+ rat retina as compared to the RCS rdy– p+ rat retina (either in the neural retina or in the RPE). Thus, there is no evidence proving yet that SR–BI might contribute to the phagocytosis of POS by RPE cells. This is in accordance with the essential role fulfilled by the c-mer tyrosine kinase receptor in the normal ingestion of POS by RPE cells in the RCS rdy+ congenic strain of rats as compared to the abrogation of this ingestion of POS in RCS rdy– congenic strain of rats. The essential role of c-mer in POS ingestion has been recently further emphasized by the production of c-mer knock-out mice displaying a RCS-like retinal degeneration [37]. We also detected SR–BI mRNA and proteins in retinal glial cells, such as oligodendrocytes, astrocytes, and in cells very likely to be Müller cells as strongly evoked by their retinal localization and their histological morphology. The presence of SR–BI in retinal glial cells is in accordance with the significant expression of SR–BI receptors in brain astrocytes [35]. Furthermore, brain astrocytes synthesize neurosteroids [38]. Recent results have strongly suggested that other brain glial cells and some neuronal sub-populations express both neurosteroids and SR–BI [39–43]. SR–BI direct involvement in neurosteroidogenesis remains to be substantiated. Our results show a high ocular expression of SR–BI and StAR but double staining experiments are mandatory for concluding decisively on the colocalization of these two receptors in neuronal and glial cells. Retinal ganglionic cells are known to produce P450scc [17], the enzyme that converts cholesterol into pregnenolone, and have the ability to synthesize neurosteroids. Our results suggest that SR–BI and StAR are produced in the same ocular cells. Independent SR–BI and StAR immunostaining experiments performed on adjacent ocular tissue sections strongly support the existence of a genuine colocalization of these two proteins in the same ocular cells. It must be stressed that the expression of StAR and SR–BI is regulated by the same transcription factors (SF-1 [44], SP-1, SREBP-1a, DAX-1 and YY1) [45,46]. Our results suggest that SR–BI and StAR are involved in some forms of steroidogenesis occurring in the rat eye and possibly in retinal neurosteroidogenesis. The cerebral distribution of StAR overlaps that of P450scc [47] and 3β-HSD. This finding implies that StAR is involved in the biosynthesis of neurosteroids from cholesterol [48] in the brain. It is tempting to speculate that this is also the case in the retina. Although neurosteroids appear to be produced only in some cerebral neuronal sub-populations, StAR appears to be strongly produced in all retinal cells. The high expression of SR–BI and StAR in the eye may suggest a key role of these genes in the ocular cholesterol metabolism and strengthens the hypothesis of the SR–BI involvement in ocular membranes biosynthesis and neurosteroidogenesis.
Acknowledgements
We are grateful to Retina France for their permanent support of CERTO. We thank Université René Descartes and the ‘Faculté de médecine Necker Enfants Malades’ for generously helping our team. We thank the ‘Ministère de la Recherche et de l'Enseignement supérieur’, INSERM, CNRS, FRM and the Fondation de France.


