1 Introduction
Post-transcriptional regulations can operate at many steps between transcription and translation, and can involve a variety of molecular mechanisms such as nuclear messenger RNA (mRNA) processing, transport of mRNA from the nucleus to the cytoplasm, regulation of mRNA stability, localisation and ability of these molecules to be translated in the cytoplasm (for a review, see [2,3]). The study of these mechanisms is greatly facilitated by the use of specialized biological systems in which no transcription takes place. In numerous animal species and particularly in amphibian, early development is programmed by maternally inherited mRNAs, which are gradually degraded during the first cleavages of the egg (for a review, see [4]). Transcription of the zygotic genome itself occurs only after the mid blastula transition (MBT) [5]. For a long time, amphibian oocyte or egg has provided an interesting experimental system for studying RNA localization [6] and translational control [7]. More recently, at the post-transcriptional level, the amphibian oocyte has also been used for mRNA nuclear export and mRNA stability analysis [8,9].
We previously set up an original in vivo system allowing the investigation of the relationship between maternal mRNA stability/degradation and appearance of zygotic transcription at MBT [10,11]. The originality of our approach was based on the use of two amphibian species, Xenopus laevis and axolotl (Ambystoma mexicanum). The axolotl egg was used as a host cell to analyse the stability/degradation of exogenously injected Xenopus RNA. The phylogenetic distance between the anurans and the urodeles allows detection of the exogenous injected transcripts without cross-hybridization to the homologous endogenous axolotl RNA.
We initially investigated the stability of the Xenopus c-myc mRNA molecules in the axolotl context. We observed that, after injection into axolotl fertilized eggs, the level of the X. laevis c-myc RNA did not decrease monotonously over time. Unexpectedly, apparent increases in RNA level could be observed at some time points [1]. To investigate this phenomenon, 35S-labelled X. laevis c-myc RNA was injected. The radioactive molecules were progressively degraded during axolotl cleavages, whereas an increase in total X. laevis c-myc RNA molecules was detected by a 32P-labelled probe. These results indicated a synthesis of c-myc RNA molecules, suggesting the possibility of a novel post-transcriptional RNA amplification process during early development.
Since this phenomenon was detected before MBT, it could correspond to a mechanism involving the machinery controlling the post-transcriptional regulation of maternal mRNA. We therefore chose to further test this hypothesis during oogenesis rather than in fertilized eggs. We show that the phenomenon is observable at different stages of oogenesis and with a wide variety of RNA template. It is sensitive to an RNA chain terminator such as cordycepin, indicating the existence of an enzymatic mechanism able to perform de novo RNA synthesis during amphibian oogenesis.
2 Materials and methods
2.1 Collection of oocytes and unfertilized eggs (UFE)
Experiments were performed on axolotl (Ambystoma mexicanum Shaw) oocytes staged according to the classification of Beetschen and Gautier [12]. Stage-VI oocytes (600–2200 μm in diameter) were isolated from ovaries, defolliculated with forceps and kept at 18–20 °C in MBSH (Modified Barth Solution High-Salt) [13]. Natural UFE were obtained from females stimulated 24 h earlier by 500 UI of chorionic gonadotropin hormone (hCG, Chorulon, Intervet, France). Maturation was induced in vitro by incubating stage-VI oocytes in 1 μM progesterone (Sigma) in MBSH for 12 h and only oocytes presenting a white area around the first polar body at the animal pole were selected (UFE Pg). Xenopus laevis stage-VI oocytes were collected, defolliculated, sized [14] and maintained in MBSH.
2.2 Preparation of synthetic RNAs and microinjection into oocytes and UFE
Capped RNAs were generated using an in vitro transcription kit (Ambion Inc., Austin, Texas, USA). Plasmids were linearised by restriction enzyme digest and proteinase K (1 U) was added for 15 min at 37 °C. They were purified by two phenol/chloroform and one chloroform extractions before ethanol precipitation. After the transcription reaction, removal of cDNA template (0.5 μg) was done by addition of RNase-free DNase (2 U) for 30 min at 37 °C. All transcripts were resuspended in Gurdon's injection buffer (88 mM Nacl, 15 mM HEPES, 1 mM KCl, 15 mM Tris/HCl, pH 7.6). From the pXLmyc plasmid containing the 2.1 kb clone of the X. laevis c-myc cDNA [15], a full-length 2.1 kb sense RNA was produced by T7 RNA polymerase after HindIII digestion. Two other X. laevis transcripts were synthesized from the c-myc X. laevis 3′UTR: after subcloning of a XhoI-BamHI fragment from pXLmyc into pBluescript KS+ (Stratagene), 0.7 kb RNAs were produced by T7 RNA polymerase after HindIII digestion (sense RNA) or by T3 RNA polymerase after XhoI digestion (antisense RNA).
The Awnt-1, Awnt5A and Awnt5B axolotl RNAs were synthesized in vitro from full-length cDNA clones ending with 8–10 adenosine residues [16,17]. The Awnt-1 clone was linearised by PstI or XbaI to generate a full-length 2.7 kb sense transcript by T3 RNA polymerase. After linearisation by KpnI, this clone generated a 2.7 kb T7 antisense transcript. From the mouse wnt1 (Mwnt1) cDNA [18], a full-length 1.9 kb sense RNA was produced by XbaI digestion and SP6 RNA polymerase. A full-length 1.9 kb in vitro transcript was produced from the PFRA clone after XbaI digestion and T3 RNA polymerase [19]. For injection, the standard protocol was as follows: 60 axolotl stage-VI oocytes or 60 UFE were microinjected in MBSH with 2 ng per cell of one synthetic transcript (corresponding to approximately 1% of the total mRNA amount present in one oocyte or UFE) in a volume of 80 nl. The time necessary to inject a transcript into these 60 cells varied between 10 to 20 min. Just after injection, all cells were pooled and a batch of 10 oocytes or UFE was taken randomly, immediately frozen at −80 °C and was labelled t=15 min. In some experiments, a batch of 10 oocytes or UFE was treated separately, injected in less than 2 min and immediately frozen (labelled t=2 min). The remaining oocytes or UFE were maintained in MBSH. Ten oocytes or UFE were taken randomly at each time point. The same procedure was applied for injection of X. laevis oocytes, except that 1 ng of the transcript was injected in a volume of 40 nl. Cordycepin 5′-triphosphate (Sigma) was co-injected with the transcripts at a final concentration of 0.25 or 2.5 mM.
2.3 RNA extraction, Northern analysis
Total RNA from each batch of oocytes or UFE was extracted using the LiCl method [20]. For Northern analysis, total RNA was electrophoresed in 1.1% formaldehyde agarose gels and transferred to Hybond N+ membranes (Amersham) [21]. Prehybridization and hybridization were done at 65 °C in Church buffer [22]. Each blot was first hybridised to the cDNA corresponding to the injected RNAs, washed at 65 °C in 2X SSC, 0.1% SDS then in 0.2X SSC, 0.1% SDS. Filters were then re-hybridised with a 32P-labelled 220 bp axolotl 18S probe and radioactivity quantified with a PhosphoImager (Amersham, Molecular Dynamics).
2.4 RT–PCR and RNase protection assays
For RT–PCR, all synthetic or total embryonic RNA samples were treated by RQ1 DNase (2 U) (Promega, France) for 30 min at 37 °C, purified by phenol/chloroform extraction, ethanol precipitated and dissolved in water. Primers used and RT–PCR conditions were previously described for Awnt-1 [16]. For RNase protection assays, synthetic sense/antisense or total RNAs were hybridised with 0.5 ng 32P labelled riboprobe in 80% formamide, 40 mM PIPES pH 6.7, 0.4 M NaCl, 1 mM EDTA, overnight at 65 °C. Non-hybridised RNAs were digested with RNases A (0.01 μg ml−1) and T1 (10 U ml−1) in 0.3 M NaCl, 0.01 M Tris-HCl pH 8.5, 5 mM EDTA for 1 h at 37 °C. Digestion was stopped by incubation with proteinase K (2 mg ml−1), 8% SDS during 15 min at 37 °C. After phenol/chloroform extraction, RNAs were ethanol precipitated and dissolved in 80% formamide, 50 mM Tris-HCl pH 8.3, 1 mM EDTA, 1% bromophenol blue, 0.1% xylene cyanol. They were electrophoresed on 5% urea-polyacrylamide gel for 1 h 30 at 300 V in TBE buffer (0.89 M Tris, 0.88 M Borate, 10 mM EDTA pH 8). For detection of the sense and antisense Awnt-1 transcript, the 426 bp AccI fragment (position 1578–2004 on cDNA) was subcloned in pSP73 plasmid. After digestion by XhoI and Sp6 polymerization, a 520 base antisense riboprobe protected a 426 base sense RNA fragment. After digestion by NdeI and T7 polymerization, a 612 base sense riboprobe protected the 426 base antisense RNA fragment.
3 Results
3.1 Fluctuations in RNA level occur in axolotl oocytes
We had previously reported that, following their injection into axolotl fertilized embryos, Xenopus laevis c-myc transcripts did not continuously decrease in abundance. Thus, over a time course, transitory increases in the amount of X. laevis RNAs were observed on a background of an overall degradation [1]. To study the developmental stage permissivity of this phenomenon, the fate of in vitro synthesized transcripts was examined after injection into axolotl stage-VI oocytes as well as in vitro progesterone matured stage-VI oocytes (UFE Pg) and in vivo unfertilized eggs obtained after stimulation of a female with hCG (UFE).
We first used as a substrate the axolotl Awnt-1 RNA, the product of a member of the wnt gene family involved in signalling pathways during early development. Sense transcripts were synthesized in vitro using as a template a 2.7 kb cDNA containing the entire coding sequence and the complete 3′ untranslated region [16]. After injection, the fate of the full-length Awnt-1 RNAs was monitored by Northern blot (Fig. 1A). Maternal Awnt-1 mRNA are present in the oocyte [23], but were below the threshold of detection of these experiments (see lane NI). In stage-VI oocytes, the Awnt-1 signal decreased between 1 and 3 h, while a 2.2-fold increase was observed between the 3- and 6-h time points. A similar pattern was detected in UFE Pg with a 2.4-fold increase between 3 and 6 h. In UFE, two increases were observed: one between 2 h 30 and 4 h (2.2 fold) and a second between 6 and 7 h 30 (1.8 fold). Similar increases were observed using a different RNA extraction protocol [24]. In addition, since the Awnt-1 signal was slightly higher (6 h post injection in UFE Pg) than in the immediately frozen axolotl oocytes (15-min time point in UFE Pg), the number of Awnt-1 RNA molecules in these samples exceeded what had been initially injected. Such an ‘overshoot’ was observed in approximately 10% of the experiments, implying that an increase in the molecules number could correspond to the observed fluctuations. As the carry over from the in vitro transcription reaction of the template and the phage polymerases could potentially result in the synthesis of additional transcripts, great care was taken to digest away the plasmid and to deproteinize the injected RNA. Control experiments (Fig. 1B) showed that no template plasmid could be detected in 20 ng of in vitro transcripts (10 times the amount of injected RNA) even after 40 PCR cycles. Thus, the injection in oocytes or unfertilized eggs of an in vitro synthesized transcript did not result in regular degradation kinetics but rather in an irregular pattern, in which some time points were more intense than the preceding ones.
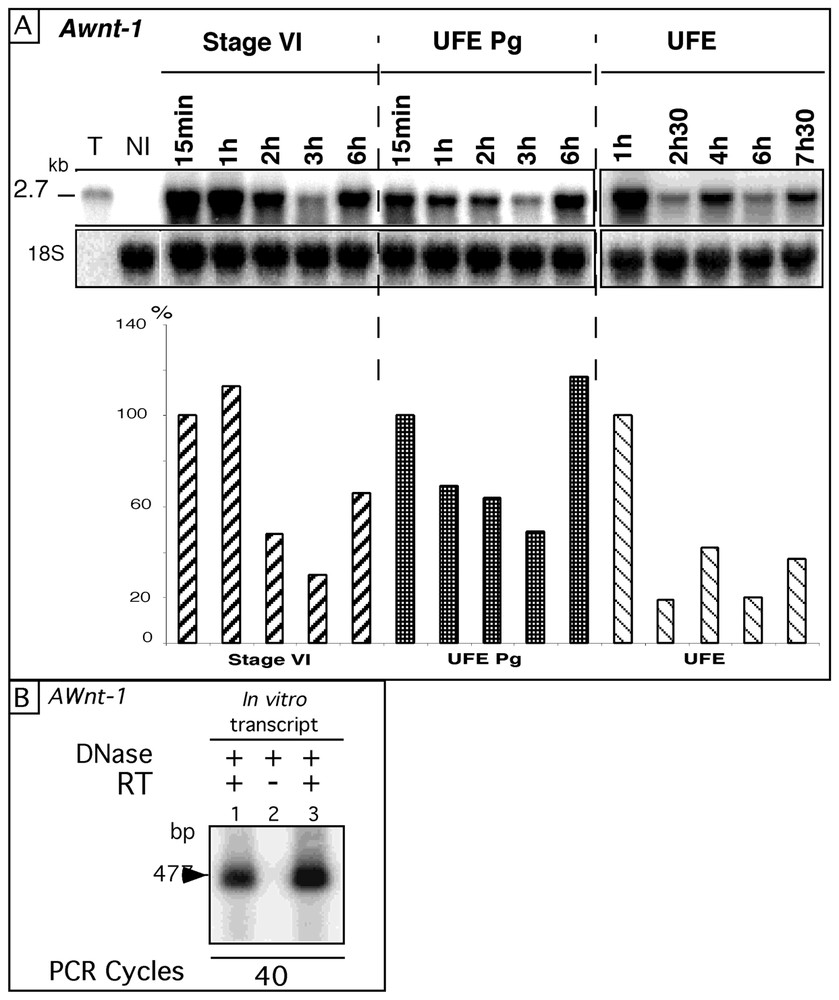
A. Fate of Awnt-1 RNA after injection into different oocyte stages. The Awnt-1 full-length sense RNA (2.7 kb) was injected into axolotl stage-VI oocytes, UFE Pg and natural UFE. Total RNA was extracted from batches of 10 oocytes or UFE taken randomly at different times after injection (15 min to 7 h 30) and 15 μg were analysed by Northern blot, hybridised successively with the axolotl Awnt-1 and the 18S cDNA probes. T: 2 ng of in vitro transcript (amount injected into one oocyte). NI: total RNA from non-injected oocytes. The histograms show Awnt-1 signals quantified with a PhosphoImager and the ImageQuant software and normalized with the 18S rRNA signal. Awnt-1 RNA levels were expressed as a percentage (%) of the t=15 min (stage-VI and UFE Pg) or 1 h (UFE) values. B. Absence of cDNA template plasmid in the in vitro transcript. 2 ng (lane 1) or 20 ng (lanes 2 and 3) of synthetic AWnt-1 RNA were treated (+) by RQ1 DNase (2U) for 30 min at 37 °C as described in § Materials and methods. Samples were assayed by RT–PCR, for 40 PCR cycles with (+) or without (−) reverse transcriptase (RT). The Awnt-1 amplified cDNA fragment is 477-base-pair long [23]. Masquer
A. Fate of Awnt-1 RNA after injection into different oocyte stages. The Awnt-1 full-length sense RNA (2.7 kb) was injected into axolotl stage-VI oocytes, UFE Pg and natural UFE. Total RNA was extracted from batches of 10 oocytes or UFE ... Lire la suite
3.2 Fluctuations in RNA level occur with several substrates as well as in Xenopus oocytes
To investigate the role of the injected RNA sequences in the observed fluctuations, we first analysed the behaviour of other full-length sense RNAs such as the 1.9-kb mouse wnt-1 (Mwnt-1) and 2.1-kb X. laevis c-myc RNAs. In these two cases, the transcripts were in the sense orientation and nearly full-length, containing the entire coding sequence of the protein as well as parts of the 5′ and/or 3′ UTR regions. As for Awnt-1, fluctuations were observed with Mwnt-1 (Fig. 2A). Control experiments confirmed that the observed signals were due to RNA molecules: pre-treatment by RNases A and T1 of total RNA from injected oocytes abolished the signals, while a RNase-free DNase pre-treatment had no effect. Two successive increases were also detected for the X. laevis RNA (Fig. 2B): a first one between 15 min and 3 h and a second one between 6 and 13 h post-injection. Similar observations were obtained after injection of the full-length 2.7-kb Awnt-5B and 3.7-kb Awnt-5A RNAs (data not shown). This suggests that the length of the injected molecule has no incidence on the fluctuation process. In addition fluctuations were observed when full-length transcripts were synthesized from cDNAs digested by different enzymes generating 5′ or 3′ protruding ends (see Table 1), indicating that the observed RNA level increase was independent of the restriction enzyme used for in vitro transcription. Thus, fluctuations could be observed with several sense transcripts in addition to Awnt-1 RNA.
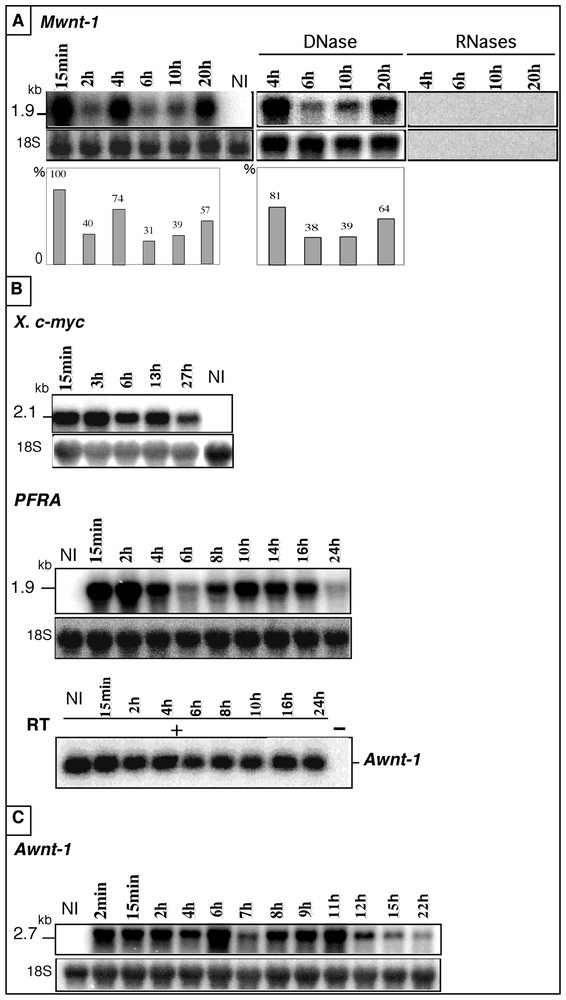
Fate of heterologous RNAs injected either into axolotl UFE or into Xenopus stage-VI oocytes. A. The 1.9-kb mouse wnt-1 (Mwnt-1) RNA was injected into axolotl UFE and total RNA was extracted from batches of 10 UFE taken randomly at different times post-injection (15 min to 20 h), analysed (10 μg) as in Fig. 1A, then hybridised successively with the Mwnt-1 and the axolotl 18S cDNA probes. The histograms show Mwnt-1 signals normalized with the 18S rRNA signal. B. The 2.1-kb X. laevis c-myc and the 1.9-kb Trypanosoma brucei PFRA RNAs were injected into axolotl UFE and analysed as in A, then hybridised successively with the radio-labelled X. laevis c-myc or PFRA cDNA probes. After injection of the PFRA RNA, the level of endogenous Awnt-1 mRNA was assayed by RT–PCR, at each time point of the experiment. 15 PCR cycles were done with (+) or without (−) reverse transcriptase (RT). NI: total RNA from axolotl non-injected UFE. C. RNA level increase of the 2.7-kb Awnt-1 RNA injected into Xenopus stage-VI oocytes. Total RNA (10 μg) extracted from batches of 10 oocytes taken randomly at different times after injection (2 min to 22 h) was hybridised with the Awnt-1 cDNA and analysed as in (A). NI: total RNA from X. laevis non-injected oocytes. Masquer
Fate of heterologous RNAs injected either into axolotl UFE or into Xenopus stage-VI oocytes. A. The 1.9-kb mouse wnt-1 (Mwnt-1) RNA was injected into axolotl UFE and total RNA was extracted from batches of 10 UFE taken randomly ... Lire la suite
Summary of the injection series. The bold numbers indicate the total number of series performed with the sense or antisense (AS) in vitro transcripts. RNA from axolotl (Awnt-1, Awnt-5A and Awnt-5B), mouse (Mwnt-1), Xenopus laevis (c-myc full-length and 3′UTR) and Trypanosoma brucei (PFRA) were synthesized after linearisation of their respective cDNAs by enzymes generating 3′ or 5′ protruding ends. The numbers between brackets indicate the number of experiments in which fluctuations(s) in the RNA level were observed
The results presented so far were obtained using full-length molecules containing the entire coding region with (Awnt) or without (Mwnt and X. laevis c-myc) complete 3′UTR, in sense orientation. Although these data indicated that the exogenous molecules could belong to phylogenetically distant vertebrate species, the synthetic transcripts used so far shared some homology with the axolotl genome. To determine if fluctuations could also be observed after injection of molecules completely unrelated to the axolotl genome, we analysed the PFRA RNA. The corresponding gene encodes the major protein A of the Trypanosoma brucei flagellum [19]. The 1.9-kb sense RNA transcribed in vitro from the complete PFRA cDNA was injected into axolotl UFE and its behaviour was examined by Northern analysis (Fig. 2B). A marked increase (3.6 fold) was observed between 6 and 10 h. Thus, fluctuations in the RNA level did not depend upon the existence of sequence similarities with the axolotl genome. In addition, fluctuations were also observed after injection of antisense PFRA RNA, indicating that the phenomenon is not restricted to sense transcripts (data not shown). These results indicate that increased RNA level can occur after injection of sense or antisense RNA templates, irrespective of the polarity of the injected transcript. In all our analyses, we used the 18S rRNA as an internal standard. To exclude that the observed fluctuations reflected a global perturbation of mRNA metabolism, we used the endogenous Awnt-1 mRNA level as an internal control after injection of PFRA sense transcript (Fig. 2B). At each time point of the PFRA time course experiment, Awnt-1 RNA level was measured by a RT–PCR assay previously used for the analysis of Awnt-1 expression during the early axolotl development [23]. No significant change in the level of the endogenous Awnt-1 transcript was observed during the 24 h time course, indicating that injection of an in vitro synthesized transcript did not globally perturb the mRNA metabolism of the recipient oocyte.
To investigate whether these fluctuations in RNA level were specific to the axolotl context or could be observed in other amphibian species, the Awnt-1 2.7-kb sense RNA was injected into X. laevis stage-VI oocytes (Fig. 2C). Note that, under our experimental conditions, the Awnt-1 probe did not detect the corresponding Xenopus transcript (non injected sample, lane NI). In this experiment, a three-fold increase was observed between 4 and 6 h and a 3.8 fold one between 7 and 11 h. This experiment indicated that the fluctuations in RNA level occur in two amphibian species, suggesting an evolutionary-conserved mechanism.
3.3 Polarity of the newly synthesized RNA molecules
As the probes used in the Northern blots presented so far were derived from double stranded cDNA, the detected signals could be due to RNA molecules of either the same or the opposite polarity as the injected ones. To clarify this, we performed in parallel a Northern blot and an RNase protection assay on RNA samples from oocytes injected with Awnt-1 sense RNA. By Northern analysis, three significant increases were observed between 10 min and 2 h, 4 and 6 h, 8 and 10 h (Fig. 3A). The RNase protection assay performed on the same samples with a single stranded riboprobe complementary to the injected transcript revealed the same pattern (Fig. 3B). Quantification of both experiments indicated an excellent agreement between the two determinations of RNA levels at all time points (Fig. 3, histogram). Similar results were observed with other substrates (data not shown). Thus, most of the signals observed in our Northern analyses were due to RNA of the same polarity as the injected transcripts.
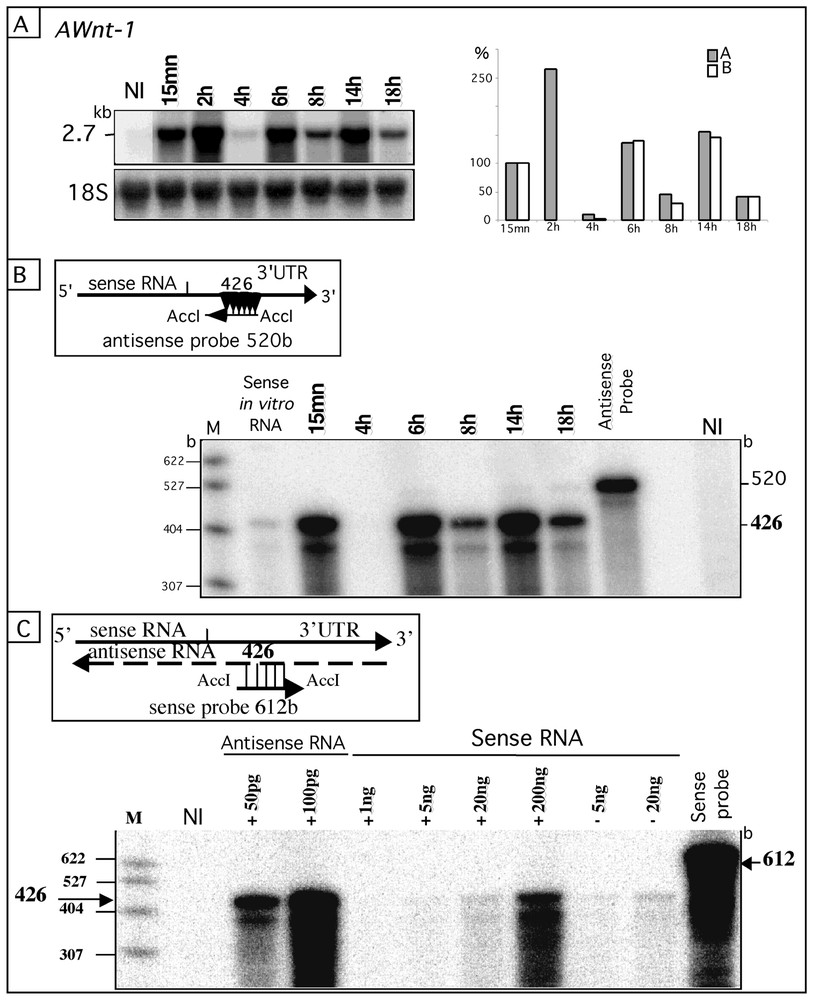
Most of the RNA molecules are of the same polarity as the injected ones, but a small amount of complementary RNA molecules are present among the in vitro synthesized transcripts. A. The 2.7-kb Awnt-1 sense transcript was injected into axolotl UFE and its abundance at different times post-injections was analysed as in Fig. 1A. The histogram (A) shows the Awnt-1 signals (grey rectangles) normalized with the 18S rRNA and expressed as a percentage (%) relative to the signal at t=15 min (100%). B. RNase protection assay. Total RNA (10 μg from the samples presented in A) was hybridised to the antisense-520-base riboprobe, which protected a 426 base fragment allowing detection of the sense Awnt-1 RNA (see diagram). These RNA signals were expressed as a percentage (%) relative to that obtained at t=15 min (hatched rectangles in the histogram). C. Detection of cRNA molecules in the injected sense transcript solution by the RNase protection assay as in (B). The 612-base-sense Awnt-1 riboprobe hybridised to the antisense transcript present in the in vitro preparation and protected the 426-base fragment (see diagram). No difference in the intensity of the 426 base fragment was observed in presence (+) or absence (−) of 20 μg of total axolotl RNA (UFE) mixed with the in vitro antisense or sense transcripts. M: pBR322-MspI digested DNA marker; NI: total RNA (10 μg) from axolotl non-injected UFE; b: base. Masquer
Most of the RNA molecules are of the same polarity as the injected ones, but a small amount of complementary RNA molecules are present among the in vitro synthesized transcripts. A. The 2.7-kb Awnt-1 sense transcript was injected into axolotl ... Lire la suite
If the increases in the RNA signals correspond to the synthesis of RNA molecules with the same polarity as the injected ones, this implies the existence of intermediate molecules for this synthesis. These intermediate molecules should be the corresponding cRNA. We searched for the presence of cRNA molecules by an RNase protection assay with a probe of the same polarity as the injected RNA. These cRNA molecules could be present initially in the injected sample and our in vitro transcripts could therefore contain trace amount of complementary RNA. Recent studies have shown that in vitro transcription by T3 or T7 polymerases contains a mixed population of sense and antisense molecules [25,26]. The amount of antisense Awnt1 cRNA molecules in the in vitro Awnt1 sense transcript sample was evaluated by RNase protection assay (Fig. 3C). The signal intensity for 200 ng of sense transcript (100 times the injected amount per UFE) was about half-equivalent to 50 pg of antisense Awnt1 RNA. This indicates that, when 2 ng of sense Awnt1 RNA were injected per UFE, 0.2 pg (10 000 fold less) antisense transcript was also injected. These results show that antisense RNA (cRNA) molecules complementary to the sense RNA are already present in trace amount in the initial sample.
3.4 Fluctuations are abolished by 3′dATP
In order to examine whether the increase in RNA signal required a de novo synthesis, exogenous transcripts were co-injected with cordycepin 5′-triphosphate (3′ deoxyadenosine (3′dATP) 5′ triphosphate), which acts as a RNA chain terminator. We analysed the behaviour of the X. laevis c-myc sense 3′UTR transcripts co-injected with the drug into axolotl UFE (Fig. 4). In the absence of the analogue, as expected, an increase of the RNA level was observed, between 3 and 4 h 30 (2.9 fold). By contrast, the co-injection of 3′dATP led to a continuous decrease of the c-myc signal. Presumably, the regular decrease in the RNA level corresponds to the degradation process of the transcript. A difference in the RNA signal intensity was registered at 15 min in the two cordycepin co-injection series (compare lanes 7 and 13). At 0.25 mM cordycepin, the RNA signal was similar to the control signal detected in absence of the drug (lane 1). At 2.5 mM cordycepin, the signal at 15 min was 30% lower than the control time point, suggesting that at a higher concentration the drug could inhibits an early wave of synthesis. Similar observations were obtained in independent additional series (data not shown). In addition to the abolition of fluctuations, the presence of an RNA synthesis inhibitor at high concentration (2.5 mM) resulted in a stabilization of the injected transcript. This stabilization was apparent even though, in the absence of inhibitor, the existence of the fluctuation phenomenon made it impossible to define a single half-life. Nonetheless, by restricting the analysis to continuously decreasing subsets of data, apparent half-lives could be obtained. Thus in this experiment, the apparent half-life of the X. laevis c-myc sense RNA was 90 min when measured between 15 min and 3 h in the absence of inhibitor and of the order of 5 h in the presence of 3′dATP (2.5 mM). Thus, inhibition of RNA synthesis had two effects on the fate of injected RNA, eliminating increases in their abundance but also suppressing an accelerated degradation. We conclude that the RNA level increase corresponding to the injected RNA molecules requires de novo synthesis.
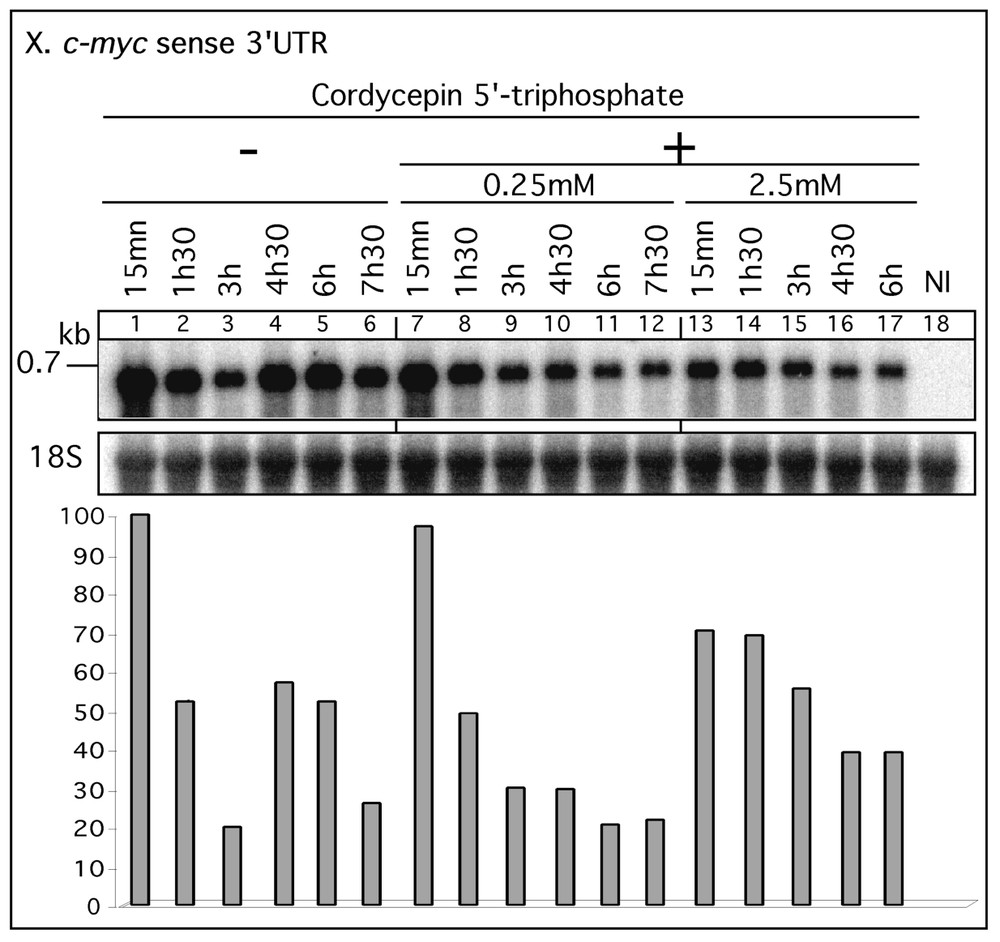
Effect of RNA synthesis inhibitor on RNA amplification. The X. laevis c-myc sense 3′UTR transcript (0.7 kb) was injected (left panel) or co-injected with 0.25 or 2.5 mM 3′dATP (cordycepin 5′ triphosphate) into axolotl UFE. Total RNA was extracted at different times post-injection (15 min to 7 h 30), analysed (10 μg) as in Fig. 1 and hybridised with the labelled c-myc 3′UTR cDNA probe. NI: total RNA from axolotl non-injected UFE. The histogram corresponds to the quantitative data of the c-myc signals normalized with the 18S rRNA and expressed as a percentage (%) of the signal at t=10 min value (lane 1). Masquer
Effect of RNA synthesis inhibitor on RNA amplification. The X. laevis c-myc sense 3′UTR transcript (0.7 kb) was injected (left panel) or co-injected with 0.25 or 2.5 mM 3′dATP (cordycepin 5′ triphosphate) into axolotl UFE. Total RNA was extracted at different times post-injection ... Lire la suite
4 Discussion
We have previously observed that, upon injection into fertilized axolotl eggs, in vitro transcribed Xenopus RNA were not degraded progressively but rather yielded an irregular pattern of decreases and increases in abundance over time [1]. Although the phenomenon was frequently observed, it was not systematic and its kinetics varied between experiments. As a prerequisite to mechanistic studies, we searched for a simpler and potentially more easily controllable experimental model by performing injection experiments into amphibian oocytes and unfertilized eggs (UFE). In the course of the present study, we performed over 50 distinct experiments (Table 1) among which 48 indicated the existence of fluctuations. Thus, this phenomenon is not restricted to the post-fertilization development of amphibians but can be detectable in stage-VI oocytes or UFE. In spite of this simpler cellular environment, the variability in the phenomenon that we had previously observed persisted throughout this study. Thus, neither the number of successive increases nor the time at which they were observed could be controlled in these experiments. Indeed, over an 18 h time course, we observed up to three successive waves of decrease and increase (Fig. 3A). Importantly, the co-injection of an RNA synthesis inhibitor such as cordycepin prevents all increases, indicating that this mechanism corresponds to a post-transcriptional RNA amplification.
4.1 Substrate and cellular specificity of amplification
We have injected RNA generated by in vitro transcription of cDNA from six different genes (Table 1). Fluctuations can be observed with a wide variety of in-vitro-synthesized transcripts, suggesting that no specific sequences are required. We have investigated the fate of both sense and antisense transcripts from the 3′UTR of the Xenopus c-myc gene and the PFRA gene. As an RNA amplification could be observed with all of these substrates, it appears that this phenomenon is not restricted to genes involved in the early development of amphibians and that neither a coding region nor a poly(A) tail are required. In addition, the size of the injected RNA is not critical as an amplification has been observed with molecules ranging in size from 0.7 to 3.7 kb.
The common point between these injected molecules is that they were all synthesized in vitrousing bacteriophage RNA polymerases. These enzymes, although highly specific, can initiate transcription at other sites than their cognate promoter [25,26]. Our molecular analysis of the in vitro synthesized RNA shows that, in addition to the ‘sense’ transcripts, trace amount of complementary transcripts (cRNA) are also present in the injection solution (Fig. 3C). In the injected solution, the cRNA molecules could therefore hybridise to the excess of sense transcript and form trace amount of double-stranded RNA (dsRNA). Therefore, the in vitro transcription could provide a key determinant for the phenomenon that we describe.
The detection of RNA amplification in oocytes and UFE, either naturally matured or in vitro matured by progesterone confirms that the underlying enzymatic machinery is of maternal origin. One common feature of these developmental stages is the extremely reduced rate of gene transcription [27], suggesting that RNA amplification is uncoupled from DNA transcription. Indeed, the fact that transcripts that are not related to any sequence present in the axolotl genome (PFRA gene of Trypanosoma brucei) can be amplified also indicates that it cannot be due to a standard transcription process. In addition, the presence of an organized nucleus is not required for amplification since it occurs in UFE in which the germinal vesicle breakdown takes place during oocyte maturation. Such RNA amplification can therefore occur in the cytoplasm as it has been already described for the de novo synthesis of globin mRNA in differentiated enucleated MEL cells [28].
4.2 Coupling between RNA amplification and degradation
Our observation of successive waves of decrease/increase in RNA level over a 18-to-24-h time course suggests that a degradation and an amplification process could act on the injected transcript. In all independent experiments, the co-injection of cordycepin (3′dATP) prevents all increases. As a RNA chain terminator, cordycepin should inhibit all classes of RNA polymerases whether DNA or RNA dependent. This demonstrates that the observed increases in RNA level require an RNA synthesis. By inhibiting RNA synthesis, cordycepin dissociate the RNA amplification from the RNA degradation process and thus allow an in vivo analysis of the ‘intrinsic’ half-life of injected RNA. In addition to preventing amplification, cordycepin also stabilizes the injected transcript, indicating that some aspect of RNA amplification plays an active role in RNA degradation. One possible mechanism could be the implication of an RNA dependent RNA polymerase (RdRP) generating dsRNA as amplification intermediates. Indeed, the presence of dsRNA can induce a specific RNA degradation mechanism [29,30]. Therefore, the observation of several cycles of amplification and degradation is very suggestive of a competition between RNA synthesis and RNA degradation.
4.3 Biological significance of RNA amplification during oogenesis and early development
Our results raise interesting issues about gene regulation during early development. In all organisms, maternal mRNAs are stockpiled and stable during oogenesis. After fertilization, their complete degradation within a few hours represents an essential developmental process allowing transcription of new messages from the zygotic genome [31]. In this study we present evidence for an RNA amplification process which could contribute both to the genesis of additional RNA molecules and to their degradation thus adding new dimensions to post-transcriptional regulations. So far, all our observations have been carried out with exogenous, in vitro synthesized transcripts containing trace amounts of complementary RNA molecules (cRNA) and thus able to form double-stranded RNA (dsRNA). It is possible that during early development some cellular mRNAs can be amplified by this process. However, we have not yet observed the amplification of an endogenous transcript. Depending on the relative activity of the amplification and degradation processes, the mechanism we have observed could play two distinct roles in gene expression. Assuming that there could be a significant delay between the amplification and the degradation phases, the amplification phase could generate additional copies of specific mRNA. Such a post-transcriptional mechanism would be particularly meaningful within the context of amphibian early development in which transcription is either severely reduced (stage-VI oocytes) or undetectable until the mid-blastula transition [32]. The observed RNA amplification/degradation phenomenon could correspond to an adaptative and conserved mechanism during evolution. Unique adaptative mechanisms exist during amphibian oogenesis such as accumulation of masked messengers and amplification of ribosomal genes [27]. Alternatively, if we assume that amplification is tightly coupled to an increased degradation, it could be a mean to degrade specific mRNA. One possible mechanism for this amplification/degradation coupling is through the induction of RNA interference (RNAi). RNAi seems to be effective in Xenopus [33–35]. We have also shown in axolotl UFE that microinjection of small interfering RNA (siRNA) directed against the endogenous Awnt-1 RNA specifically degrades these transcripts but not the Awnt-5A or Axdazl endogenous RNA (unpublished results). This strongly suggests that the RNAi machinery is effective in our biological system.
It will be of obvious interest to characterize the polymerase activity involved in this RNA amplification. We initially excluded the involvement of an RNA polymerase II since α-amanitin did not inhibit this RNA synthesis [1]. It is also of interest to note that it takes place in a context where no transcription occurs. Moreover, post-transcriptional amplification can be observed with RNA unrelated to the axolotl genome as illustrated by the PFRA gene of Trypanosoma brucei. The coupling we observed could correspond to a component of an endogenous RNA interference process implying amplification due to an RNA polymerase RNA dependent (RdRp) activity. Genetic evidence suggests that an RdRp plays a critical role in the establishment of RNA interference in plants, N. crassa and C. elegans [36–38] and Drosophila [39]. The amphibian oocyte provides an excellent experimental model to analyse such cellular activity and its biological significance.
Acknowledgements
The authors are very grateful to O. Albagli, A. Audibert, P. Bastin, L. Dandolo, A. Lefranc, P. Nardeux for helpful comments. They thank P. Bastin, M.W. King, R. Nusse and C. Séguin for providing the Trypanosoma brucei PFRA cDNA clone, the Xenopus c-myc cDNA clone, the mouse Wnt-1 cDNA clone, the Awnt1–5A and-5B cDNA clones, respectively. This work was supported by grants from the ‘Ligue régionale de Normandie’ and the ‘Ligue régionale Île-de-France contre le cancer’.


