1 Introduction
The innate immune system exists in all multicellular organisms; only vertebrates have, in addition, the adaptive immune system for fighting microbial infection. The innate system is comprised of a limited number of proteins dedicated to the recognition of signatures of microbial infection. These proteins, termed pattern recognition receptors or PRRs, are encoded by the germ-line of the animal and thus are not subject to much genetic variation [1]. Because of this, the molecular patterns that are recognized by these proteins are highly conserved structural motifs of the infecting microbe. These microbial motifs are termed pathogen associated molecular patterns or PAMPs [1]. Some examples of PAMPs are lipopolysaccharide (LPS) from the outer membrane of Gram-negative bacteria and peptidoglycan (PGN), which is found in the cell wall of both Gram-negative and Gram-positive organisms. Once detection of the PAMPs has occurred by the host PRRs, the signal is transmitted inside leading to the activation of transcription factors such as NF-κB, which drive the expression of genes whose protein products play a key role in the defense response against the invading microbe.
2 TLRs and Nods
Recent studies have suggested that two systems of microbial detection exist in the mammalian host. One system comprises a family of membrane-bound receptors called the Toll-like receptors or TLRs. TLR4, which is probably the best characterized member of this family, detects LPS present in the extracellular milieu [2]. The other family of detection proteins is the nucleotide-binding site/leucine-rich repeat (NBS/LRR) family [3,4]. The best characterized members of this family are Nod1 and Nod2. These proteins are located in the cytoplasm and are involved in the detection of bacterial PAMPs that enter into the cell either with an invasive microbe or by translocation by certain pathogenic bacteria through specialized transfer apparatuses.
Why does the host require an intracellular means of pathogen detection in addition to the TLRs? The cytoplasmic detection system mediated by Nods likely plays a key role in host defense in those tissues where TLRs are absent or expressed at low levels [5]. This occurs in epithelial cells that line mucosal surfaces, like for example, in colonic epithelial cells. Since these cells live in constant contact with the microbial flora, expression of TLRs and/or their co-factors is down-regulated, especially on the surface epithelium, in order to avoid stimulation of the cells by PAMPs and aberrant inflammation of these tissues. However, when these cells are infected with invasive pathogens or pathogens that interact intimately with the plasma membrane, PAMPs can be transferred to the intracellular compartment where they interact with Nod proteins and this then initiates the defense response [6].
The domain structure of Nod1 and Nod2, characterized by a central nucleotide binding site (NBS) and a C-terminal leucine-rich repeats domain (Fig. 1), is reminiscent of the domain organization of plant disease resistance proteins or R proteins [3,4]. R proteins in plants, for example RPS2 in Arabidopsis, are involved in sensing microbes that enter the cytoplasmic compartment and initiating a defense response. Because of this similarity with plant R proteins, Nods were hypothesized also to play a role in host defense in mammalian cells. Activation of the Nod molecules is thought to occur when a bacterial ligand is sensed by the LRR domain. Activation of the molecules then occurs leading to self-oligomerization through the NBS region. Through the N-terminal caspase-activation and recruitment domain (CARD), Nod1 and Nod2 (which has an additional CARD domain compared to Nod1) can then activate the NF-κB pathway leading to the expression of a number of pro-inflammatory genes by this transcriptional regulator (see reviews [3,4]).
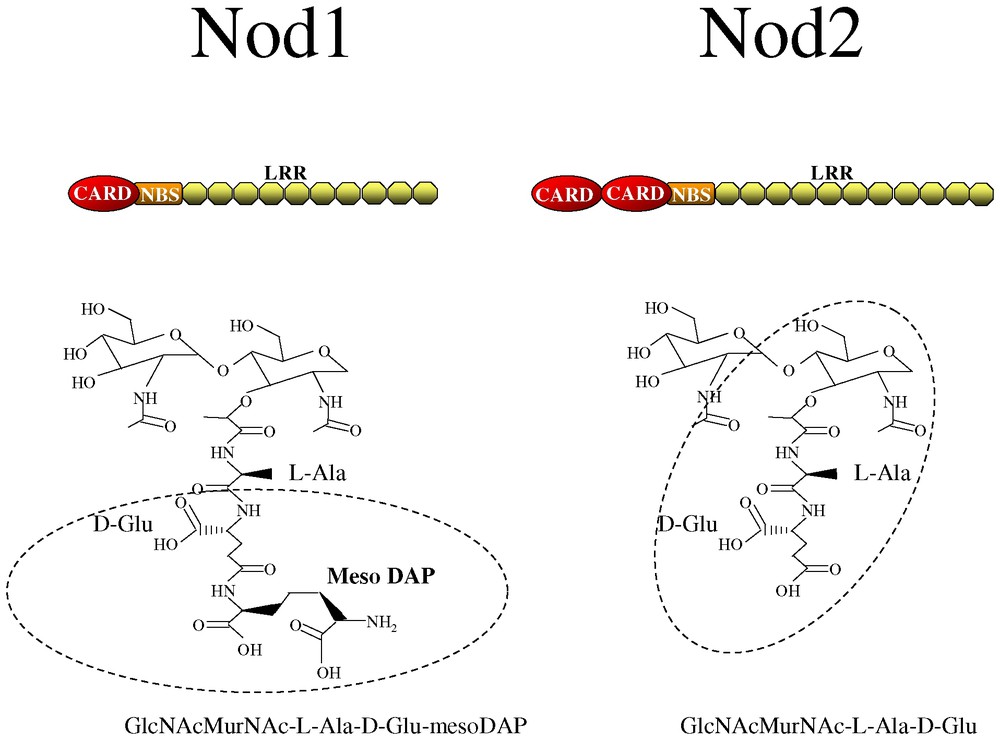
Domain structure of Nod1 and Nod2 and the naturally occurring PGN motifs recognized by these proteins, GlcNAcMurNAc-l-Ala-d-Glu-mesoDAP and GlcNAcMurNAc-l-Ala-D-Glu. The minimal motifs, d-Glu-mesoDAP and muramyl dipeptide (MurNAc-l-Ala-d-Glu) are shown in circles. CARD, caspase activation and recruitment domain; NBS, nucleotide binding domain; LRR, leucine-rich repeat.
3 Nods detect unique PGN fragments
Our recent studies focus on identifying the bacterial ligands or PAMPs that activate Nod1 and Nod2. For Nod1, we screened a number of different potential PAMPs for their ability to activate NF-κB in a Nod1-dependent manner. Strikingly, we found that purified PGN from Gram-negative bacteria including Escherichia coli and Shigella flexneri, were able to activate Nod1 leading to NF-κB induction. PGN preparations from the Gram-positive bacteria, Bacillus subtilis and Staphylococcus aureus, were unable to stimulate this response. Through the analysis of Gram-negative bacterial PGN fractions separated by reverse-phase HPLC we were then able to determine that the minimal naturally occurring PGN fragment that stimulates Nod1 is a disaccharide of N-acetyl glucosamine-N-acetyl muramic acid linked to a tripeptide where the terminal amino acid is meso-diaminopilimic acid (meso-DAP) (Fig. 1) [7]. The presence of DAP in the PGN can be considered as a general signature of Gram-negative bacterial infection since most, but not all, Gram-positive organisms have lysine in this position in their PGN. What is also striking for Nod1 is that it is highly specific for the tripeptide structure since the presence of an additional amino acid to the DAP abrogates the sensing of this bacterial product by Nod1 [7].
More recently, we and others have been able to narrow down the PGN structure sensed by Nod1 in more detail thus defining the minimal motif. Markedly, Nod1 can sense just the terminal two amino acids within the naturally occurring fragment: d-glutamine-mesoDAP (Fig. 1) (referred to as iE-DAP by the group of Inohara) [8,9]. This minimal motif is a signature of bacterial infection since neither of these amino acids exist in mammals.
In the context of in vivo infection, Nod1 appears to be a key molecule in the sensing of Gram-negative bacterial infection since isolated intestinal epithelial cells from mice deficient in Nod1 can no longer react to the intracellular presentation of Gram-negative bacterial products. Moreover, Nod1 appears to be the only functional bacterial sensor in intestinal epithelial cells highlighting the key role of this PRR in host defense [7].
As mentioned above, Nod2 is highly homologous to Nod1 except for an additional N-terminal CARD domain. The expression of profile of Nod2 is more or less restricted to monocytes/macrophages but expression can be upregulated in other cell types upon treatment with pro-inflammatory stimuli such as interferon γ or TNFα [10,11]. Nod2 has been recognized as an important mediator of inflammatory induction since the gene encoding Nod2 is the first susceptibility gene that has been identified for the chronic inflammatory bowel disease, Crohn's disease [12,13]. Our studies also led us to examine the bacterial ligand sensed by Nod2. Like Nod1, this protein also senses a PGN fragment, however, Nod2 is specific for muramyl dipeptide or MDP (Fig. 1) [14,15]. MDP is the minimal bioactive PGN fragment from both Gram-positive and Gram-negative bacteria making Nod2 a general sensor of bacterial infection. Interestingly, MDP has been known for decades for its immunomodulatory action; it is a component of Freunds complete adjuvant. With the discovery of Nod2 as the host receptor for MDP, the immunostimulatory properties of this compound can now be investigated in more detail.
4 Role of Nod2 in Crohn's disease
The most common mutation in the Nod2 gene that is associated with Crohn's disease is an insertion mutation at position 3020 that leads to the deletion of the terminal LRR of the protein. When we compared the wild-type versus the mutant protein for the ability of these molecules to sense MDP, it was observed that the mutant Nod2 can no longer detect MDP to initiate NF-κB activation [14]. Moreover, the group of Gabriel Nunez showed that peripheral blood mononuclear cells isolated from patients with Crohn's disease could not respond to MDP in terms of NF-κB activation and cytokine induction [15]. The implications of these findings therefore suggest that the defect in Crohn's disease patients may be the inability to respond normally to bacterial products. An apparent paradox thus arises: how can the lack of bacterial detection and loss of subsequent activation of pro-inflammatory signals translate into the chronic and severe inflammation of the bowel that we see during Chohn's disease? One possible explanation is that the intestinal mucosa is maintained at a certain level of inflammation by Nod2 and possibly Nod1 in order to keep in check any possible bacterial breach of the epithelial barrier. If bacteria or their products enter within the sub-mucosa, Nod2 induces a local inflammatory response that suppresses any possible infection. In Crohn's disease, it can be hypothesized that without this sentinel function of Nod2, bacteria and/or their products that gain access to the sub-mucosa over stimulate the system, aggravating defense systems that would not normally be active. This effect would manifest itself as the severe and chronic inflammation that is observed in the intestinal tissues of Crohn's disease patients.
5 Conclusion
In conclusion, Nod1 and Nod2 have now been defined as representatives of a new family of cytoplasmic pathogen recognition molecules. These two proteins recognize distinct motifs found in the PGN of bacteria and initiate defensive responses in the host through the activation of NF-κB. What will be interesting in the future will be to define the responses down-stream of Nod1 and Nod2 and to compare these responses to those down-stream of the TLRs. In this way, the specificity of responses emanating from these different PRRs can be characterized in order to better understand the individual role of these proteins in defense responses against microbial infection.
Acknowledgements
We would like to thank Stephen Girardin for comments on the manuscript and the members of the groups of ‘Immunité innée et signalisation’, ‘Pathogénie bactérienne des muqueuses’ and ‘Pathogénie microbienne moléculaire’ at the Institut Pasteur and others who contributed to the work discussed in this review.


