1 Introduction
Transcriptional regulation of mRNA-encoding genes in eukaryotes requires additional proteins beyond activators and the RNA polymerase II (RNA pol II) transcriptional machinery. These additional proteins, termed coactivators, are complex molecular machines composed of multiple subunits. The coactivators fall into one of two classes. The first class is composed of enzymes that affect chromatin structure, either through covalent modification of the histones, or by an ATP-dependent remodeling of chromatin architecture. The second class is defined principally by their interactions with RNA pol II and its associated proteins, the general transcription factors (GTFs: TFIIA, TFIIB, etc.). The proteins of this second class have collectively been termed mediators or adaptors, because they serve as a bridge between the activator and the basal transcription machinery consisting of RNA pol II and the general transcription factors. The TBP-associated factors or TAFs constitute the first class of adaptors that was discovered. TAFs have essential roles in activated but not in basal transcription. Some TAFs have been shown to contact GTFs and others contact activators. TAFs are generally thought to be in permanent association with TBP, forming TFIID, often considered one of the GTFs. However, studies in several systems suggest that TAFs play more specific roles than those played by the other GTFs. The second class of adaptors that was discovered now bears the name Mediator and is the subject of this review. Mediator appears to have a unique role in transducing the signal from a transcription factor to the transcriptional machinery.
The exact nature of the role of Mediator in RNA pol II-dependent transcription, however, is still unclear. The paradigm for Mediator function is that it recruits RNA pol II to the promoter, passively transmitting information from the activator to the transcriptional machinery. Recent data suggest a more involved and complex role for Mediator in transcription. In this model Mediator plays an active role and is itself subject to regulation. For example, Mediator could be regulated through post-translational modifications like phosphorylation. If so, then Mediator can be thought of as the ultimate regulator of transcription, combining information from activators, promoter architecture, and perhaps even directly from upstream regulatory signals. The sum of this regulatory information could then be modulated and communicated to the transcriptional machinery. In this capacity Mediator could have the ability to alter both the activation and the repression of a gene. There is emerging evidence that this is the case, and one of the focal points of this review will be a discussion of that evidence and how it supports the idea that Mediator has a negative role in transcriptional regulation.
Mediator was first identified in the yeast Saccharomyces cerevisiae, an ideal model organism for studying transcriptional regulation because of the extensive genetic, molecular and biochemical approaches that can be utilized in concert to study biological problems. This review will be largely limited to a discussion of the yeast Mediator complex and its role in both positive and negative regulation of transcription.
2 Mediator components
2.1 Identification of Mediator components
Mediator was first discovered in the yeast Saccharomyces cerevisiae in the early 1990s. The story of the identification of Mediator, from the initial concept to the final catalogue of subunits is an example of the powerful combination of genetics and biochemistry that illustrates the utility of yeast for tackling complex biological problems.
The first indication of the requirement of a coactivator came from in vitro transcription assays in which it was observed that the purified activators and transcription machinery were not sufficient for transcription, even upon addition of excess quantities of every component known to be essential for basal transcription. Activator-dependent transcription required the addition of a crude yeast fraction [1]. The unknown component that provided activator-dependence was given the name Mediator. Further purification of Mediator based on the activator-dependent transcription assay allowed the identification of some of the components in this complex fraction. The biological significance of the Mediator complex was revealed when it was found that several of the first polypeptides identified biochemically are encoded by genes associated with defects in transcriptional regulation of gene expression.
The most important of these genetic studies, carried out independently of the biochemical identification of Mediator, utilized a unique property of the RNA pol II, its repetitive carboxyl terminal domain (CTD). The CTD contains 26 or 27 repeats of a heptapeptide sequence that is conserved in eukaryotes [2]. The CTD is essential for viability, but the number of repeats can be reduced and viability is maintained. The truncation of the CTD to only 10 to 12 repeats results in a cold-sensitive growth phenotype [3]. Nonet and Young [4] took advantage of this phenotype and screened for suppressors that would restore growth in the cold, due to a gain of function mutation of a component that interacted with the CTD. They first identified a dominant mutation in the non-essential gene SRB2 (suppressor of RNA polymerase B subunit mutation). Further analysis of dominant suppressors yielded SRB4, SRB5, and SRB6 [5]. These components have since been renamed, but for historical clarity, they are referred to here by their original names. Table 1 lists the old and new names for all Mediator components, the protein module to which they belong, their approximate sizes, and summarizes some of their distinctive phenotypes.
Nomenclature of S. cerevisiae mediator components, after Bourbon et al. [15]
| Gene | Former designation(s) | Module | Size (kD) | Deletion phenotype |
| MED1 | middle | 64 | viable | |
| MED2 | tail | 48 | viable | |
| MED3 | PGD1, HRS1 | tail | 43 | viable |
| MED4 | middle | 32 | inviable | |
| MED5 | NUT1 | middle | 129 | viable |
| MED6 | head | 33 | inviable | |
| MED7 | middle | 26 | inviable | |
| MED8 | head | 25 | inviable | |
| MED9 | CSE | middle | 17 | viable |
| MED10 | NUT2 | middle | 18 | inviable |
| MED11 | head | 15 | inviable | |
| MED12 | SRB8, GIG1, SSN5, NUT6 | CDK8/CycC | 167 | viable |
| MED13 | SSN2, SRB9, UME2, NUT8, SCA1, RYE3 | CDK8/CycC | 160 | viable |
| MED14 | RGR1 | middle | 123 | inviable |
| MED15 | GAL11, RAR3, SDS4, SPT13, ABE1 | tail | 120 | viable |
| MED16 | SIN4, BEL2, GAL22, SDI3, SSF5, SSN4, TSF3, RYE1 | tail | 111 | viable (ts) |
| MED17 | SRB4 | head | 78 | inviable |
| MED18 | SRB5 | head | 34 | viable |
| MED19 | SSN7, NUT3, ROX3 | head | 25 | viable |
| MED20 | SRB2, HRS2 | head | 23 | viable |
| MED21 | SRB7 | middle | 14 | inviable |
| MED22 | SRB6 | head | 14 | inviable |
| MED23 | ||||
| MED24 | ||||
| MED25 | ||||
| MED26 | ||||
| MED27 | ||||
| MED28 | ||||
| MED29 | ||||
| MED30 | ||||
| MED31 | SOH1 | middle | 14 | viable |
| CDK8 | SRB10, SSN3, UME5, GIG2, NUT7, RYE5 | CDK8/CycC | 63 | viable |
| CycC | SRB11, SSN8, UME3, RYE2, NUT9, GIG3 | CDK8/CycC | 38 | viable |
Other Mediator components were identified by further exploitation of the CTD mutations. Additional work using the same cold-sensitive screen identified the recessive suppressors SRB7–SRB11 [6,7]. A different screen made use of the fact that a substitution of alanine or glutamate for the serine at position two or five in the heptapeptide repeat sequence of the CTD is lethal. SRB9 (MED13) was also identified in this screen as a suppressor of the substitution of alanine or glutamate at position two [8].
Of the nine SRB genes, only three are essential, SRB4 (MED17), SRB6 (MED22), and SRB7 (MED21). Deletion of the other SRB genes leads to distinct phenotypes which are described in detail elsewhere [9]. SRB10 (CDK8) and SRB11 (CycC) encode a cyclin-dependent Ser/Thr protein kinase and its associated cyclin, respectively, that are involved in negative regulation of gene expression. Their deletion leads to the up-regulation of a number of genes, as discussed in more detail later.
Other Mediator components were identified in genetic screens of a different nature, carried out over many years by investigators studying a wide variety of gene regulatory networks. This accounts for the large number of aliases by which the Mediator subunits are known, and is further confirmation of the general role that Mediator plays in the regulation of gene expression.
The identification of yeast Mediator components was largely completed by the biochemical isolation of a 20-subunit complex that was purified to homogeneity [10]. Many of the subunits identified in the complex are products of genes that were identified previously in screens for mutations affecting transcription. Seven of the purified subunits are encoded by novel genes that are named MED (for Mediator). For a more thorough discussion of the discovery of yeast Mediator components, the reviews by Myers and Kornberg [11] and Carlson [9] are recommended.
The first human Mediator complexes were discovered concurrently using in vitro activator-dependent assays [12]. Later components of the mammalian Mediator complex were identified by immunopurification assays based on homologues of yeast Med7, Med21 and Cdk8 [13]. The exact composition of mammalian Mediator is less well defined, in part due to the loss of subunits during the purification process. A recent analysis of Med8-containing rat liver Mediator by mass spectrometry attempted to clarify this issue by merging data obtained from six independent purifications, resulting in the identification of 30 distinct MED subunits [14]. Extensive analysis has shown that nearly every component of the yeast Mediator has a metazoan homolog [15]. Different components appear to be conserved to varying degrees, however, giving rise to the idea that those components which interact with RNA pol II are more highly conserved than those components that interact with the various activators, allowing Mediator to adapt to the more rapid evolution of the various promoters while still maintaining a conserved mechanism [16].
2.2 Nomenclature
One of the results of the cumulative efforts to identify Mediator components and the discovery of the conservation of the subunits throughout eukaryotes is a redundant and piecemeal nomenclature system. The yeast Mediator components are, for the most part, named to reflect the type of screen conducted to find the mutant (e.g., SSN, suppressor of Snf1; UME, unscheduled meiosis; NUT, negative regulation of URS Two, etc.). This led to multiple names for the same gene because many Mediator components have pleiotropic effects on transcription, especially mutants affecting the subunits of the tail and the middle modules. Bourbon et al. recently proposed a unified nomenclature for Mediator subunits in which all components are named MED followed by a number. The originally named MED subunits will retain their numbers (MED1–MED11), and the other subunits will be named starting with MED12, in order of decreasing molecular weight. The only exceptions are SRB10 and SRB11, which will be renamed CDK8 and CycC, respectively, due to the prevalence of this nomenclature in the mammalian literature for this CDK-cyclin pair [15]. Table 1 lists the original names along with the new names.
2.3 Organization and structure
A combination of biochemical, genetic and structural data suggest that the yeast Mediator is composed of three functionally and physically distinct modules or subcomplexes (Fig. 1). The head module, composed of Med6, Med8, Med11, Med17, Med18, Med19, Med20, and Med22, which can be isolated intact biochemically, is thought to have a general role in transcription and interacts with the CTD of RNA pol II [17].
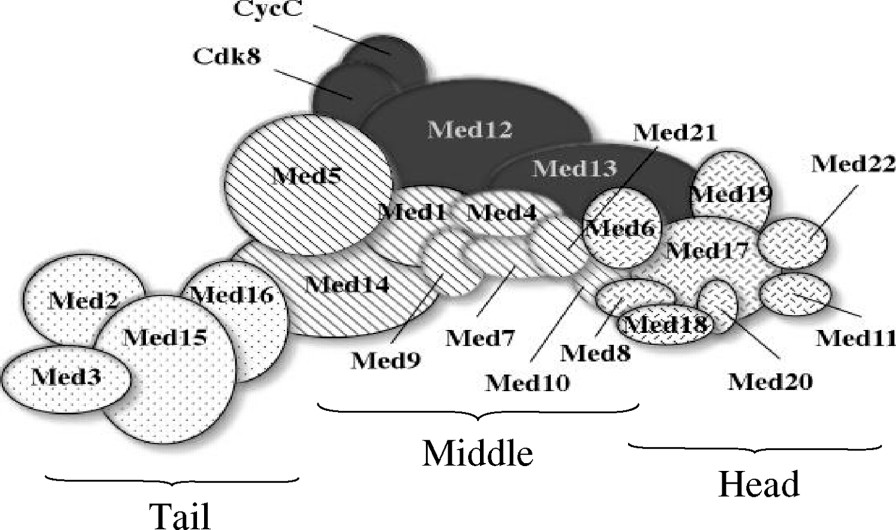
Cartoon of Mediator after Guglielmi et al. [19] showing the approximate sizes and positions of components. Positions are intended to reflect the current data regarding interactions of the Med proteins.
The second module, the middle domain, which is composed of Med1, Med4, Med5, Med7, Med9, Med10, Med14, and Med21 is believed to be a subcomplex based on its de novo reconstitution from recombinant proteins [18]. There is some debate about whether Med14 is part of the middle domain or part of the tail due to its unique position, which connects these two domains. In a recent paper describing the interactions of Mediator components, Guglielmi et al. placed Med14 in the tail [19], whereas Dotson et al. refer to the middle domain as the Rgr1 (Med14) module in their structural work [20]. It is undisputed, however, that Med14 is the bridge from tail to middle. Like the head, the middle interacts with the CTD of RNA pol II. In addition, the general transcription factor TFIIE interacts with the middle module.
The evidence for the tail module, composed of Med2, Med3, Med15, and Med16 is largely genetic. Deletion of any one of the members of this module leads to a similar phenotype [21,22]. This module is presumably responsible for recognizing and binding to activators [23].
There may be a fourth distinct subcomplex composed of the Med12, Med13, Cdk8, and CycC subunits. This presumed module contains the cyclin-kinase pair Cdk8 and CycC which phosphorylates the CTD at the serine 5 residue in the heptapeptide repeat [24]. Loss of function mutations in either Cdk8 or CycC lead to the derepression of a subset of genes [25], suggesting a role for these subunits in repression.
The best structural information available on yeast Mediator is a ∼35-Å resolution three-dimensional structure that was generated from electron-microscope images [26]. These image reveal two conformational states of Mediator: a compact arrangement seen in the absence of RNA pol II, and a second elongated structure consisting of three domains observed in the presence of RNA pol II. The three domains are named according to their position relative to RNA pol II: the head makes contacts with RNA pol II, the middle, and the tail, which makes contacts with activators.
This elongated structure is only seen in the presence of RNA pol II. However, if the CTD is truncated by removing the heptapeptide repeats, the Mediator remains in the elongated structure [26]. This supports the idea that Mediator is making contact with more than just the CTD of RNA pol II. This was the first evidence that Mediator makes multiple contacts with RNA pol II, some of which involve the middle domain of Mediator as well as the head domain. In contrast to the structure seen in the presence of RNA pol II, Mediator maintains the compact form in the presence of the activator GAL4-VP16 [26]. Binding to RNA pol II or to the activator has different effects on the structure of Mediator, suggesting that the order of binding may influence function.
It is a natural suggestion that, given the myriad screens in which Mediator components were identified, the individual components themselves may have functions outside of their roles in the Mediator complex. To date, however, there is little evidence that this is true. Extensive tap-tagging projects carried out by Greenblatt and co-workers failed to find Mediator components in any other complexes (personal communication). It is possible, however, that under some conditions subunits or modules of Mediator could be present independent of the intact complex [27].
In addition to yeast Mediator, structures have also been generated for both murine and human Mediator [20]. The structures of all three complexes appear to be conserved at the 30–40-Å resolution range. This overall structural similarity fits well with the homology of the individual components observed between yeast and higher eukaryotes and emphasizes the universal nature of Mediator.
3 The role of Mediator
3.1 Global regulation
The widespread importance of Mediator is obvious from genome-wide expression analysis. Holstege et al. performed genome-wide expression assays with a variety of Mediator and RNA pol II mutants. In an Rpb1 ts mutant, they found that nearly all mRNAs decreased in comparison to the wild type strain. They take this level to be the baseline for RNA pol II dependent transcription and use that standard for comparison. A similar analysis in a Med17 ts strain shows that the level of expression of 93% of genes affected by a ts mutation in Rpb1 is also decreased by more than two-fold [28]. Loss of other components has a less dramatic effect. For example, the same microarray analysis showed that a Med6 ts mutant affects only 10% of the genes compared to the loss of Rpb1. A point mutation in Cdk8 that renders it catalytically inactive was also used in this assay, and this Cdk8 mutant showed an increase in expression of 173 genes (about 3% of the whole genome), indicating the repressive role of Cdk8. These results point to the fact that there are some gene- or activator-specific aspects of Mediator's role, but it is clear that nearly all activator-dependent transcription requires at least some components of Mediator.
In addition to playing a role in activator-dependent transcription, there is also evidence that Mediator is required for basal transcription. It was found that basal transcription in a crude yeast extract was dependent on Med20 and Med18, but that this requirement was alleviated when purified components were used [5,10,29]. A similar pattern of dependence was also found for RNA pol II CTD [30], suggesting that basal transcription is dependent on Mediator to overcome a non-specific repressor, and that Mediator acts on the CTD in order to do so.
The role of Mediator as an interface between gene-specific regulatory factors and the general transcriptional machinery allows it to act as a global regulator of transcription. Mediator is essential for transcription at nearly every promoter in yeast, which supports the idea that it is Mediator rather than an activator that is recognized by RNA pol II. In this sense, it could act to mask individual promoters and present them all in a uniform manner to RNA pol II. This fits nicely with the above expression data to create a model in which some components interact with RNA pol II, while others recognize activators.
Another way in which Mediator is a global regulator is in its dual ability to act as both a coactivator and a corepressor. The majority of findings point to Mediator's role as a coactivator, but there is also strong evidence, including the identification of some Mediator components based on increased rather than decreased levels of transcription in their absence, that suggests a negative role for Mediator in transcription as well, although that effect may be indirect, as discussed later.
3.2 Function and mechanism
An intriguing possibility explaining the mechanism of Mediator is that it first serves as a repository for information from activators, promoter architecture, and perhaps even up-stream regulatory signals acting directly on it, and then serves as a modulator of that information, passing it on to RNA pol II in a controlled manner. This could offer several advantages: first that RNA pol II is always receiving the signal from the same complex, instead of from various activators, and secondly, it allows for many levels of control, including negative regulation. Although the exact mechanism of Mediator-dependent activated transcription is not yet clearly defined, there have been many studies that aim to elucidate the nature of its role in this process.
Recruitment of RNA pol II to the preinitiation complex (PIC) was the earliest idea put forth to address the function of Mediator. In this model, the tail domain would recognize and bind to activators, leading to the subsequent recruitment by the head domain of RNA pol II. The recruitment model is supported by experiments in which the LexA DNA binding domain was fused to the Mediator components Med15, Med16 and Med19 (Med 15 and 16 are both in the tail domain; Med19 is in the head domain). The protein fusions strongly activated transcription from a reporter under the control of a promoter containing LexA binding sites [31–33].
This simple idea, that Mediator recruits RNA pol II to the PIC, was complicated by the finding that ‘recruitment’ entails different things at different promoters, especially in regards to artificial activation [34]. The promoter architecture (position and number of activation domains, chromatin state) affects the ability of an activator to recruit RNA pol II to the PIC, suggesting that the involvement of Mediator is more than just a physical bridge spanning the gap between activator and CTD. It implies that Mediator (or activators exerting their effect through Mediator) has roles beyond recruitment of RNA pol II.
A plausible manner in which Mediator could be playing such a secondary role is through chromatin remodeling. Mediator contains histone–acetyltransferase (HAT) activity [35]. HATs acetylate nucleosomes, an activity which is generally associated with a loosening of DNA and consequent increased access of the transcriptional machinery to the promoter. It has been shown that Mediator can interact directly with nucleosomes [35], and furthermore, that it contains Med5, the subunit responsible for the HAT activity. Med5 is non-essential, but it may be that Mediator-dependent chromatin remodeling is not needed at any essential promoters, or another chromatin remodeling complex could compensate for the loss of Med5. Yet another possibility is that under some conditions, Mediator recruits a separate chromatin remodeling complex such as Gcn5 to the promoter.
It has been found that the role of Mediator extends beyond the formation of the PIC. Spurred on by the observation that actively transcribing polymerase lacks associated Mediator, Svejstrup et al. [36] proposed that Mediator is involved in a cycle of association to form the PIC and then dissociation as RNA pol II begins elongation, followed by reformation of the PIC for a second round of transcription. This cycle parallels the phosphorylation and dephosphorylation cycle that the CTD undergoes. One of the components of the Mediator, Cdk8, stimulates CTD phosphorylation by TFIIH [10]. It was later shown that Mediator remains at the promoter after transcription initiation [37]. Taken together, these results suggest a role for Mediator as a scaffold for PIC formation and a possible role in the CTD phosphorylation cycle.
Continuing along the same line of logic, that Mediator ultimately exerts its effect directly on RNA pol II, Mediator is hypothesized to serve as a gateway for RNA pol II to access the promoter. The limiting factor in the rate of transcription is the number of molecules of RNA pol II that can enter the transcription cycle in a given amount of time. Using this as a guideline, Lewis and Reinberg [38] proposed a model in which the ultimate function of Mediator is to provide RNA pol II with varying levels of access to the promoter, corresponding to the appropriate regulatory environment. According to this model, the regulation is achieved largely through promoter architecture. The number and spacing of upstream regulatory elements as well as the state of the chromatin would define the structure that Mediator assumed, which would in turn define the rate of transcription.
3.3 Mediator as a corepressor
A paradoxical aspect of Mediator that remains unresolved is its apparent ability to play both a positive and a negative role. The first indication that Mediator could act as a repressor came from a screen in which a recessive mutation in MED16 allowed expression of HO in the absence of the Swi5 transcriptional activator [39]. Since then, many other examples of the repressive effect of Mediator have been found, but there is no consensus mechanism for it. Indeed, it is not yet clear if the effect is direct or indirect.
In the original identification of Mediator subunits, mutants of MED15 were isolated that both activated transcription inappropriately, or were partially defective for transcription under the appropriate physiological conditions [9]. Immunoprecipitation experiments have shown that Med15 forms a subcomplex with Med16 and Med3 (tail module) as well as with Med14, the bridge between the tail and middle [17,40]. Nishizawa et al. tried to reconcile the fact that Med15 has both a negative and a positive role in transcription by suggesting that Med15 acts as an activator when it is unable to form that subcomplex, but assumes a negative role when the subcomplex is intact [41]. However, since a deletion of MED15 has the same phenotype as the MED15-ts allele used by Nishizawa, an alternative explanation is that the tail subcomplex lacking Med15 acts in a negative manner. Med16 is known to have a general repressive role [32] at many promoters. The mechanism by which its presence causes a switch in the role of Med15, however, is unclear.
In general, whenever the structure of the tail domain of Mediator is disrupted by mutating one of its subunits, or when its association with the middle domain is altered by disrupting the function of Med14, repression of many genes is alleviated. Thus the structural integrity of the tail domain appears to be important for appropriate gene expression. In particular, an intact tail domain is necessary to prevent expression of genes that are in a non-expressed state because of the inactivity or absence of their activators, or because they lack a UAS sequence. However, gene expression in these circumstances still required an appropriate TATA sequence in the case of one artificial promoter tested [41]. In this model, the make-up of the Mediator complex determines if it acts as a repressor or activator.
Post-translational modifications of specific Mediator subunits have also been suggested to account for Mediator's ability to switch between a positive and a negative regulator. Changes in gene expression of the FLP genes on the yeast two micron plasmid result from Cdk8-dependent phosphorylation of the Med2 subunit [42]. No chromosomal genes were detectably affected when the Med2 phosphorylation sites were mutated to Ala. Nevertheless, this result shows that Mediator function can be affected by post-translational modification of a single Mediator subunit, suggesting the possibility that Mediator itself is regulated.
Yet another proposed mechanism is that Mediator exerts its negative effect indirectly by recruiting a general corepressor, the Cyc8-Tup1 complex [43]. This corepressor does not bind directly to DNA, but rather is recruited to the promoter by DNA-binding repressor proteins such as Mig1, Rox1, Ume6 and Mcml/α2 and al/α2. A physical interaction detected by two-hybrid and GST pull-down assays between the Mediator subunit Med3 and this corepressor supports the idea that Mediator is acting indirectly to repress transcription. If this were the case, a mutation in CYC8 or TUP1 should have the same phenotype as a mutation in a Mediator component that relieves repression. This was shown not to be the case for one artificial promoter studied, [41] suggesting that this model does not apply universally. In a related set of experiments it was shown that mutations in the MIG1 and MIG2 repressor genes acted synergistically with mutations in MED16 and MED14, suggesting that these Mediator components do not act through the Cyc8-Tup1 complex [44]. Another possibility for an indirect repressive role of Mediator is its effect on the transcription of the corepressor Cyc8-Tup1 and the DNA binding proteins that recruit it. Genome-wide microarray expression data show however that loss of Med17, Med18, Med6 or Med10 does not lead to any obvious change in the transcription levels of those genes [28]. This evidence suggests that Mediator's repressive role may sometimes by indirect, but that is not always the case.
As noted above, one of the functions of Mediator is to stimulate phosphorylation of the CTD. The CTD is initially unphosphorylated when bound by Mediator, and then hyperphosphorylated while free from Mediator during elongation. It has been suggested that this activity is an avenue for negative regulation. In such cases RNA pol II would be unable to form a stable PIC because Mediator would stimulate phosphorylation of the CTD prematurely, causing RNA pol II to dissociate [36]. This model implies that the formation of a stable PIC requires interactions following the arrival of RNA pol II before the PIC is fully functional. By causing premature phosphorylation of the CTD, Mediator could prevent stable PICs from forming.
There is some evidence that contradicts a direct role of Mediator in repression, or even an indirect role that acts though a corepressor. In a study of GAL genes, it was shown by chromatin immunoprecipitation that the Mediator components Med14, Med5, Med7 and Med22 are not found at promoters of inactive genes [45]. This would suggest that Mediator is only playing a negative role indirectly. It is possible, however, that Mediator was not detectable at inactive promoters because it is either weakly or transiently associated with inactive promoters. The evidence that Mediator plays a negative role at GAL genes is based on the activity of these genes that is observed when subunits of the tail module are absent. In this case the effect of the Mediator mutations could be indirect. For example, loss of Med15 function could reduce the level of the Mig1 repressor that is able to recruit the Tup1 repressor complex to the GAL promoters. This hypothesis is consistent with the evidence demonstrating that the same ts allele of MED15 produces phenotypes suggestive of both positive (failure to grow on galactose) and negative (relief from repression) affects of Med15 on gene expression.
A similar explanation could explain the effect of mutations in MED14 and MED16 on chromatin structure. Jiang and Stillman [21] demonstrated a change in linking number, and hence presumably nucleosome density, on episomal plasmids when the plasmids were present in strains with defects in MED14 or MED16. They suggested that these genes might alter gene regulation through effects on chromatin structure. Similar arguments were advanced to explain the increased accessibility of an episomal plasmid to dam methylase expressed in yeast with defective MED15 or MED16 genes [41]. However, defects in these Mediator components could lead to reduced levels of a histone chaperone activity necessary for nucleosome assembly. These experiments draw attention to the fact that it is not known definitively if the repressive effect of Mediator is direct or indirect.
4 Current directions and perspectives
The emerging picture of human Mediator, which echoes that of yeast Mediator, proves the universal nature of the coactivator as a regulator of all RNA pol II eukaryotic transcription. It also demonstrates the power of using yeast as a model organism to tease apart the details of a complex system. Regardless of the system being used, however, there is still much left unknown, which precludes our understanding of the finer workings of this large, multi-functional complex.
High-resolution structural information is still lacking, and it is likely to become available only in small pieces due to the limitations of current technology to resolve such large structures. New technology using photo-crosslinking to map protein-protein interactions could provide key information about the structure of Mediator.
The intriguing possibility of Mediator itself being regulated suggests that it may function as a ‘checkpoint’ for transcription initiation, preventing RNA pol II from gaining access to the template under inappropriate conditions, or serving as a scaffold to facilitate RNA pol II binding and transcription initiation. Promoter architecture, other coactivators and corepressors, as well as other upstream regulatory signals would all influence whether RNA pol II would form a functional PIC, or whether it would dissociate because Mediator was in a repressive state. This model fits much of the experimental data, and also correlates with other examples of cellular checkpoints. It also provides a rationale for such a large, complex molecular machine as the intermediary between activators and RNA pol II because it would have to sense not only the activator but also the state of the chromatin and other regulatory factors.


