1 Introduction
The order Squamata includes lizards (ca. 4750 sp.), snakes (ca. 3000 sp.), and amphisbaenians (ca. 160 sp.). Together with the two extant species of tuataras from New Zealand, they form the Lepidosauria [1–5]. Squamates are divided into two major clades based on morphology: the Iguania (Iguanidae, Agamidae, Chamaeleonidae) and the Scleroglossa (Dibamidae, Amphisbaenia, Serpentes, Gekkonidae, Xantusiidae, Lacertidae, Teiidae, Gymnophthalmidae, Scincidae, Cordylidae, Anguidae, Xenosauridae, Shinisauridae, Helodermatidae, Varanidae). Additionally, the Autarchoglossa include all scleroglossans except the gekkonids, with the dibamids and amphisbaenians generally considered as Scleroglossa incertae sedis [6–15].
According to this arrangement, which has been widely accepted, a major event in squamate evolution was the switch from tongue prehension of food used by the muscular-tongued tuataras and iguanians to the teeth and jaw prehension of prey used by the hard (keratinized) tongued scleroglossans, freeing the tongue for chemoreception. This presumably allowed the scleroglossans to exploit a variety of habitats and foraging modes unavailable to iguanians and to dominate in squamate assemblages throughout the world [16–21].
Although other higher-level squamate relationships are still controversial, the following groupings are supported by most morphological studies: Acrodonta (Agamidae, Chamaeleonidae), Teiioidea (Teiidae, Gymnophthalmidae), Lacertiformes (Lacertidae, Teiioidea), Scincoidea (Scincidae, Cordylidae), and Anguimorpha (Anguidae, Xenosauridae, Shinisauridae, Helodermatidae, Varanidae, Serpentes) (see reviews by Evans [2] and Lee et al. [20]).
Most previous molecular studies of squamates have not sampled a diversity of taxa and thus conclusions have been limited. In early 2004, the first molecular study to sample all major squamate lineages [22], including all lizard and amphisbaenian families and most snake families, discovered some unconventional relationships among squamates. With sequences of two nuclear genes (C-mos and RAG1), snakes were found not to be nested within Anguimorpha. In addition, a close relationship was found between lacertid lizards and amphisbaenians, and between those taxa and the teiioid lizards. The classical association of snakes with either the varanid lizards or amphisbaenians was therefore rejected. Moreover, the classical squamate dichotomy between Iguania and Scleroglossa was not supported. Instead, Iguania (one fifth of all living species of squamates) was found to be a derived lineage, clustering high in the tree, together with snakes and anguimorphs. Later in 2004, a second molecular study [23] reached similar conclusions with additional sequence data and support. Nonetheless, several major nodes in both studies were weakly supported or unresolved.
In this work, we expand upon those earlier studies with new sequences from additional genes in an attempt to obtain a more robust phylogeny of squamate reptiles. Here we present analyses of sequence data from nine nuclear protein coding genes (C-mos, RAG1, RAG2, R35, HOXA13, JUN, MAFB, α-enolase, and amelogenin) obtained from all major squamate lineages. It is the first study to use the JUN, MAFB, and amelogenin genes to elucidate squamate phylogenetic relationships, and expands the taxonomic coverage of other genes (RAG2, R35, and HOXA13).
2 Materials and methods
2.1 Taxonomic sampling and DNA sequencing
Tissue samples were obtained from the tissue collections of Nicolas Vidal and S. Blair Hedges (see Appendix A). We avoided taxa characterized by an increased rate of evolution in order to decrease the influence of long-branch attraction artifacts. Therefore, within snakes, we used henophidian representatives, which evolve more slowly than scolecophidians and caenophidians [24]. For the same reason, within Iguania, we used iguanid representatives, which evolve more slowly than Acrodonta (Chamaeleonidae and Agamidae) [22,23]. DNA extraction was performed using the DNeasy Tissue Kit (Qiagen). The sets of primers used for amplification and sequencing are listed in Appendix B.
Both strands of the PCR products were sequenced using the BigDye sequencing kit (Applied Biosystems) in the ABI Prism 3100-Avant Genetic Analyser. The two strands obtained for each sequence were aligned using the BioEdit Sequence Alignment Editor program [25]. Accession numbers of sequence data obtained from GenBank are listed in Appendix C. The new sequences have been deposited in GenBank.
2.2 Sequence analysis
Sequence entry and alignment (19 taxa) were performed manually with the MUST2000 software [26]. Amino acid properties were used, and ambiguous gaps deleted. This resulted in 360 C-mos sites, 723 RAG2 sites, 732 R35 sites, 444 HOXA13 sites, 330 JUN sites, 81 α-enolase sites, 336 amelogenin sites, and 324 MAFB sites. The RAG1 dataset (2862 sites) was obtained from Townsend et al. [23]. In all analyses, gaps were treated as missing data.
We built phylogenies using probabilistic approaches, with Maximum-Likelihood (ML) and Bayesian methods of inference. ML analyses were performed with PAUP*4 [27]. Bayesian analyses were performed with MrBayes 3.0b4 [28,29]. For ML methods, an appropriate model of sequence evolution was inferred using ModelTest [30], both for separate and combined analyses. As we used only nuclear protein coding genes, and because separate analyses showed no significant topological incongruence, we performed combined analyses, which are considered to be our best estimates of phylogeny. For the concatenated dataset (6192 sites, 3217 variable sites), the model selected is the model (base frequencies: A (0.2804), C (0.2548), G (0.2262), T (0.2386); substitution parameters: A-G: 3.8461, C-T: 4.3629; proportion of invariable sites (I): 0.3326; gamma distribution shape parameter (G): 1.6449). Bayesian analyses were run with model parameters estimated as part of the Bayesian analyses, and the best-fit model as inferred by ModelTest. ML results are presented under the form of a bootstrap majority rule consensus tree, which is considered to be a reliable estimate of phylogeny [31] (1000 replicates; NJ starting tree with NNI branch swapping). Bayesian analyses were performed by running 2 000 000 generations in four chains, saving the current tree every 100 generations. The last 18 000 trees were used to construct a 50% majority-rule consensus tree.
We estimated dates for lineage divergences using the Bayesian dating method implemented in Multidivtime [32–34]. Model parameters were estimated from the concatenated data set using PAML [35]. The MCMC parameters were 10 000 MC samples, 100 cycles, and 100 000 burnin cycles. The following five fossil-based calibration points were used as minimum constraints: oldest anguimorph (Parviraptor, Bathonian, 166 Myr) [2], oldest helodermatid (Primaderma, Albian, 106 Myr) [36], oldest teiid (Ptilotodon, Aptian-Albian, 112 Myr) [37], oldest cordylid (Konkasaurus, Maastrichtian, 68 Myr) [38], and oldest rhineurid (Plesiorhineura, Danian, 64 Myr) [39]. We set the maximum age of squamates at the Permian/Triassic boundary (251 Myr) (RTTM) based on the fossil-based inference by Evans [2] (origin of squamates at 240–230 Myr). The resulting dates are presented in Fig. 1 and differ by no more than about 5% from dates (unrelated to the maximum age constraint itself) estimated in separate analyses (1) by setting the maximum age of squamates at the Jurassic/Triassic boundary (200 Myr) or (2) by constraining the maximum age of anguimorphs at 166 Myr.
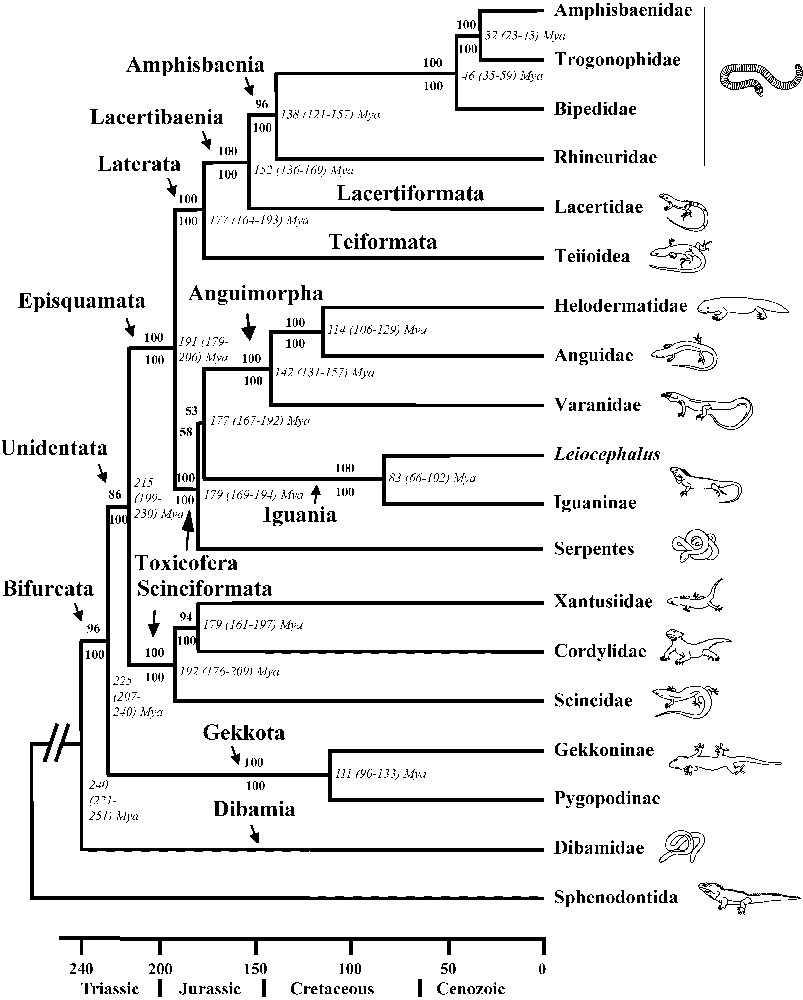
Phylogenetic relationships of squamates inferred from DNA sequences of nine nuclear protein-coding genes (C-mos, RAG1, RAG2, R35, HOXA13, JUN, α-enolase, amelogenin, and MAFB, totaling 6192 bp). Branch lengths are proportional to time. Values in bold above branches are ML bootstrap values. Values in bold below branches are Bayesian posterior probabilities. Values in italics next to nodes are time estimates (followed by error range).
3 Results and discussion
3.1 Higher-level squamate relationships
Our nuclear dataset allows us to resolve with strong support all major squamate splits but one (Fig. 1). The limbless dibamids are the most basal lineage, followed by the gekkonids. All squamates but Dibamidae have a variously bifurcated tongue (from slightly notched in gekkonids to deeply forked in varanids and snakes) [7,16,40]. The only other exception lies within Chamaeleonidae, which have lost the bifurcated condition and have secondarily acquired a highly specialized tongue that is projected to capture prey [7,16]. As the tongue of Sphenodon is not notched, the presence of a bifurcated tongue is therefore a shared derived trait uniting the squamates composing the closest relative of dibamids (Fig. 2).
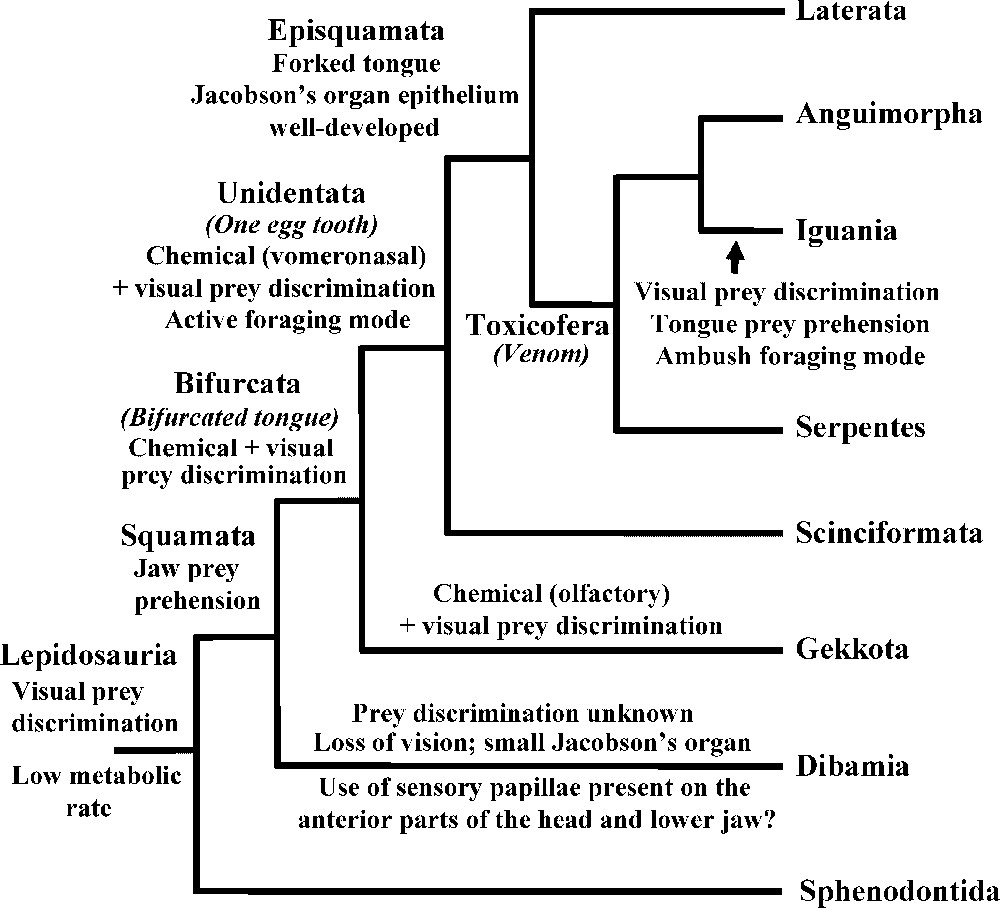
Evolution of prey discrimination and prehension in squamates. Squamates relied primitively on visual prey discrimination, jaw prey prehension and a low metabolic rate. Chemical prey discrimination (whether olfactory or vomeronasal) evolved with Bifurcata (bifurcated tongue). Gekkota rely mostly on olfactory and visual prey discrimination, and retain a relatively low metabolic rate (mostly ambush foragers). Unidentata are more active foragers (higher metabolic rate) and rely primarily on vomeronasal and visual prey discrimination. The vomeronasal prey discrimination culminates with Episquamata, which includes the squamates with the most deeply bifurcated tongues, best-developed Jacobson's organs, and the highest metabolic rates. Iguanians display several reversals in these traits.
Dibamids and gekkonids are the only two squamate lineages possessing paired egg teeth [40,41]. The paired egg teeth condition is therefore primitive for squamates, and the presence of a single median egg tooth is a shared derived trait uniting all squamates composing the closest relative of gekkotans (Fig. 2).
The next higher-level group comprises xantusiids, cordylids, and scincids, with scincids in a basal position, which is consistent with previous molecular studies [22,23,42]. This group is the closest relative of a large clade that includes other lizards as well as amphisbaenians and snakes, and which is divided into two major groups. The first group comprises amphisbaenians, lacertids, and teiioids; within this clade, the teiioids are the most basal lineage, and amphisbaenians and lacertids form a monophyletic group, all results previously found by Vidal and Hedges [22], Townsend et al. [23], and Fry et al. [43]. Within amphisbaenians, rhineurids are the most basal lineage, followed by bipedids, which are the closest relatives to a clade including amphisbaenids and trogonophids, a result in accordance with previous molecular studies [22,23,43–45]. The second major group comprises iguanians, anguimorphs, and snakes, with snakes as the most basal lineage, although this last result is not strongly supported. The presence of toxin secreting oral glands is a shared derived trait of this clade, demonstrating a single early origin of the venom system in squamates, instead of two independent origins (one among caenophidian snakes and one among helodermatid anguimorphans) [43] (Fig. 2). Within anguimorphs, varanids are in a basal position, as found by Vidal and Hedges [22], Townsend et al. [23], and Fry et al. [43].
The position of snakes among squamates has been a controversial topic. The closest relatives of snakes are neither the varanids, nor burrowing lineages such as amphisbaenians and dibamids (which are limbless or have reduced limbs), but are the anguimorphs and/or the iguanians. The novelty of this phylogenetic arrangement mainly lies in the clustering of iguanians with anguimorphs and snakes [43]. Our study therefore rejects the monophyly of both Scleroglossa and Autarchoglossa, because iguanians are in a highly nested position among Squamata. The two lepidosaurian lineages using tongue prehension of food, the tuataras and the iguanians, have therefore acquired their feeding modes independently. As the iguanians are the only squamate lineage using tongue prehension of food, and are highly nested within squamates, we can robustly infer that they have lost the jaw prehension trait used by all other squamate lineages, and have secondarily acquired their tongue prehension trait, an inference also made by Townsend et al. [23] (Fig. 2). In parallel, iguanians have switched from visual and vomeronasal prey discrimination to visual (only) prey discrimination (Fig. 2). Iguanians thus do not represent a primitive state of evolution in squamates but are a species-rich (ca. 1440 sp.) and specialized lineage combining lingual prehension, dependence on visual cues, and ambush foraging mode, and which feeds mainly on low-energy and noxious prey avoided by other squamates such as ants, other hymenopterans, and beetles [21]. Given this type of food, the functional significance (e.g., in defense) of the toxin secreting mandibular and maxillary glands present in iguanians [43] remains to be investigated.
3.2 Taxonomic implications
Our proposed taxonomic changes are indicated in Fig. 1. Names are not required for every node of a phylogeny, but squamates are widely used in ecological and evolutionary studies, and higher-level taxonomic names such as these facilitate research and communication in the field. For taxonomic stability, we prefer to use available names. However, when the definition (characters) and content of some taxa are altered substantially, it is less confusing and more practical to abandon those old names and use new names, as we have done in several cases here. In each case, the taxon named refers to the most recent common ancestor of the included groups and all descendants.
Lacertiformata (new name) includes Lacertidae. Lacertibaenia (new name) includes Lacertiformata and Amphisbaenia, and is a name that includes elements of both words. Teiformata (new name) includes Teiidae and Gymnophthalmidae. Although one solution to the problem of the paraphyly of Lacertiformes would be to redefine this taxon to include Amphisbaenia, this would conflict with the meaning of the name (‘lizard-shaped’) and lead to confusion over content. Therefore, we propose a new name, defined by a morphological character that is consistent with the molecular phylogeny. Laterata (new name) includes Lacertibaenia and Teiformata, referring to the presence of tile-like (squarish or quadrangular, and sometimes raised) scales that form the rings in Amphisbaenia, and are also present ventrally in Lacertiformata and Teiformata (while recognizing that squarish scales occur in other taxa, such as xantusiids and some anguimorphs). Toxicofera (new name) includes Iguania, Anguimorpha, and Serpentes, referring to the presence of venom. Episquamata (new name) includes Toxicofera and Laterata, referring to its derived position in the tree of squamates (‘top squamates’). Scinciformata (new name) includes Scincidae, Xantusiidae, and Cordylidae. Although one solution to the problem of the paraphyly of Autarchoglossa would be to redefine this taxon to include Iguania, this would conflict with the meaning of the name (‘free-tongued’) and lead to confusion over content. Our new name, Unidentata, includes Episquamata and Scinciformata and refers to the presence of one egg tooth. Bifurcata (new name) includes Unidentata and Gekkota, referring to the presence of a bifurcated tongue. With the repositioning of Iguania to a derived position in the tree, the name ‘Scleroglossa’ now becomes obsolete in terms of its meaning and content and is discarded.
Our proposed classification of Squamata is summarized here:
| Squamata |
| Dibamia |
| Bifurcata |
| Gekkota |
| Unidentata |
| Scinciformata |
| Episquamata |
| Toxicofera |
| Serpentes |
| Unnamed clade |
| Anguimorpha |
| Iguania |
| Laterata |
| Teiformata |
| Lacertibaenia |
| Lacertiformata |
| Amphisbaenia |
3.3 Biogeography
The new relationships and divergence times provide a better understanding of the historical biogeography of squamates. Molecular time estimates show that the Triassic and Jurassic (from 250 to 150 Myr) were important times for squamate evolution and diversification (Fig. 1). For example, the early divergences among the major groups (Dibamia/Bifurcata, Gekkota/Unidentata, Scinciformata/Episquamata) apparently took place largely during the Triassic at a time when all of the continents were joined in a single supercontinent Pangaea. Therefore, we should not expect a strong geographic influence in the ancestral distributions of these groups. However, the major clades of episquamatans contain lineages that diverged during the Jurassic and later, after the initial breakup of Pangaea, and therefore we should expect to see more geographic patterns in their historical (e.g., Mesozoic) distributions (Cenozoic distributions show considerable dispersal among continents). For some groups, such as anguimorphs (Laurasia), snakes (possibly Gondwana) and iguanians (possibly Gondwana), this has already been noted [2]. The association of those three groups in Toxicofera now raises the possibility that vicariance has played a role in their origin. Another striking pattern in the tree (Fig. 1) is the relatively recent (Cenozoic) origin of three lineages (families) of amphisbaenians: Bipedidae, Trogonophidae, and Amphisbaenidae. Currently, the major limitation of drawing biogeographic inferences is the relatively poor fossil record of squamates from the southern hemisphere [2]. Nonetheless, this new molecular phylogeny and timescale of squamate evolution should help encourage development of new hypotheses and searches for new fossils.
Acknowledgements
We thank those persons and institutions who helped us and contributed tissue samples used in this study: A. Bauer, R. Bezy, B. Branch, S. Busack, J. Campbell, C. Cicero, K. Daoues, C. Daugherty, B. Demeter, H. Dessauer, A. Dubois, B.G. Fry, C. Gans (supported by Leo Lesser Foundation), C. Hass, R. Highton, V. Koyamba, G. Lecointre, Louisiana State University Museum of Zoology, L. Maxson, A. Miralles, P. Moler, T. Moncuit, P. Moret, T. Pappenfus, A. Ross, J.-Y. Sire, S. Sweet, University of California Museum of Vertebrate Zoology (Berkeley), and A. Wynn. We especially thank A.-S. Delmas for her invaluable assistance in the laboratory, F. Battistuzzi for help with some analyses, J.-C. Rage for help with paleontological issues, and R. Bour and P. David for help with taxonomic issues. This work was funded by the ‘Service de systématique moléculaire’ of the ‘Muséum national d'histoire naturelle, Institut de systématique’, Paris (CNRS FR 1541) to N.V. and by grants from the NASA Astrobiology Institute and National Science Foundation to S.B.H.
Appendix A Tissue samples used
The following list identifies the collection number of the sample used and the locality. Collection numbers have the following prefixes: LACM (Los Angeles County Museum), MVZ (University of California Museum of Vertebrate Zoology, Berkeley), NV (Nicolas Vidal, ethanol preserved tissue collection), SBH (S. Blair Hedges, frozen tissue collection), and USNM (United States National Museum, Smithsonian, Washington, D.C.).
Sphenodontida: Sphenodon punctatus (SBH 266085; New Zealand; RAG2, R35, HOXA13, JUN, amelogenin), Squamata: Xantusiidae: Xantusia vigilis (LACM 136813; California, San Bernadino County, Hesperia; RAG2), Xantusia henshawi (LACM 136789; California, Riverside County, 4.7 miles SE Banning; R35), Xantusia riversiana (LACM 125513; California, Ventura County, San Nicolas Island; HOXA13), Cricosaura typica (SBH 190532; Cuba, Santiago de Cuba Province, 2.8 km N. Uvero; JUN, MAFB, amelogenin), Amphisbaenidae: Amphisbaena cubana (SBH 161956; Cuba, Guantánamo Bay Naval Station; RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase), Rhineuridae: Rhineura floridana (SBH 172913; Florida, Plant City; RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase), Bipedidae: Bipes biporus (MVZ 137543; Baja California Sur; RAG2, HOXA13), Bipes canaliculatus (SBH 267134; Guerrero, Mexico; R35, JUN, MAFB, amelogenin, α-enolase), Trogonophidae: Trogonophis wiegmanni (MVZ 11124; North Africa; RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase), Lacertidae: Podarcis hispanica (SBH 266666; Morocco, Asilah; RAG2, JUN), Takydromus sexlineatus (NV; locality unknown; R35, HOXA13, MAFB, amelogenin), Teiioidea: Teiidae: Ameiva auberi (USNM 306540; Cuba, Guantánamo, 8.9 km SW Hatibonico; RAG2, HOXA13, JUN, MAFB), Gymnophthalmidae: Gymnophthalmus underwoodi (SBH 102274; Guadeloupe, Grande Terre, Playa Anse Laborde; amelogenin), Iguanidae: Cyclura nubila (SBH 104677; Puerto Rico; C-mos, RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase), Leiocephalus barahonensis (SBH 101427; Dominican Republic, Barahona Province, Barahona; C-mos, RAG2, R35, HOXA13, JUN, MAFB, amelogenin), Anolis sagrei (SBH 160990; Jamaica, St James Province, 3.2 km W. Mocho; α-enolase), Gekkonidae: Gekkoninae: Gekko vittatus (NV; Indonesia; RAG2, R35, HOXA13, JUN, MAFB, amelogenin); Pygopodinae: Lialis burtonis (SBH 266057; Papua New Guinea, Wipim; RAG2, R35, MAFB, amelogenin), Lialis jicari (NV, Irian Jaya, JUN), Cordylidae: Cordylus giganteus (SBH 266055; South Africa; RAG2, R35, HOXA13, JUN, MAFB, amelogenin), Scincidae: Eumeces inexpectatus (SBH 191579; Florida; RAG2, R35, JUN, MAFB, amelogenin), Helodermatidae: Heloderma suspectum (SBH 194118; locality unknown; RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase), Dibamidae: Dibamus novaeguinea (SBH 266054; Philippines; RAG2, R35, HOXA13, JUN, MAFB, amelogenin), Anguidae: Anniella pulchra (SBH 194106; California, Santa Barbara County, Santa Maria; RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase), Varanidae: Varanus dumerilii (SBH 266058; locality unknown; RAG2, R35, JUN, MAFB, amelogenin, α-enolase), Serpentes: Pythonidae: Liasis savuensis (NV; Savu Island, Indonesia; RAG2, R35, HOXA13, JUN, MAFB, amelogenin, α-enolase).
Appendix B Primers used
Amplification was performed using the following sets of primers: G73, 5′-GCG-GTA-AAG-CAG-GTG-AAG-AAA-3′ [46], G74, 5′-TGA-GCA-TCC-AAA-GTC-TCC-AAT-C-3′ [46] for the C-mos gene; L460, 5′-AAC-AAT-GAN-CTT-TCT-GAT-AA-3′ (original), L562, 5′-CCT-RAD-GCC-AGA-TAT-GGY-CAT-AC-3′ (original) and H1306, 5′-GHG-AAY-TCC-TCT-GAR-TCT-TC-3′ (original) for the RAG2 gene; L29, 5′-CTG-AAA-ATK-CAG-AAC-AAA-A-3′ (original), L29B, 5′-CTG-AAA-ATG-CAG-AAC-AAA-AGT-AC-3′ (original), L42, 5′-GAA-CAA-AAG-TAC-WGT-TTC-AAT-3′ (original), L75, 5′-TCT-AAG-TGT-GGA-TGA-TYT-GAT-3′ (original), H786, 5′-TTG-GRA-GCC-ARA-GAA-TGA-CTT-3′ (original), H792, 5′-CAT-CAT-TGG-RAG-CCA-AAG-AA-3′ (original), H792B, 5′-CAT-CAT-TGG-GAG-CCA-RAG-AAT-GA-3′ (original) for the R35 gene; F2, 5′-ATC-GAG-CCC-ACC-GTC-ATG-TTT-CTC-TAC-GAC-3′ [47], F35, 5′-GTC-ATG-TTY-CTY-TAC-GAC-AAC-AG-3′ (original), F54, 5′-ACA-ACA-GCY-TGG-ARG-AGA-TYA-ACA-A-3′ (original), R2, 5′-TGG-TAG-AAA-GCA-AAC-TCC-TTG-3′ [47], R2B, 5′-TGG-TAG-AAA-GCA-AAC-TCC-TTG-G-3′ (original), R2*, 5′-GCC-CTG-GTA-GAA-RGC-RAA-CTC-CT (original) for the HOXA13 gene; L731, 5′-TGG-ACT-TCA-AAT-CCC-CCG-ATG-ATC-CCA-GC-3′ [42], H912, 5′-CCA-GGC-ACC-CCA-GTC-TAC-CTG-GTC-AAA-3′ [42] for the α-enolase gene; LJUN, 5′-CAG-TTC-YTS-TGC-CCC-AAG-AA-3′, LJUN2, 5′-CCA-AGA-ATG-TCA-CYG-AYG-AGC-A-3′, HJUN, 5′-GAC-TCC-ATG-TCR-ATR-GGG-GA-3′, HJUN2, 5′-GGA-GGA-GTC-TCC-CCA-GGC-ATT-T-3′ for the JUN gene; LMAF2, 5′-TSG-AGG-AYC-TGT-ACT-GGA-TG-3′, HMAF, 5′-CAC-CTC-RTC-YTT-GGT-GAA-GCC-3′ for the MAFB gene; LAM2D, 5′-TAY-CCA-CRK-TAY-DSY-TAT-GAR-CC-3′, LAM2N, 5′-TAT-CCA-CGT-TAT-GGC-TAT-GAA-CC-3′, HAM, 5′-CAC-TTC-YTC-YTK-CTT-GGT-YT-3′ for the amelogenin gene.
Appendix C Sequence data obtained from GenBank
Sphenodontida: Sphenodon punctatus (RAG1: AY662576; C-mos: AF039483), Squamata: Xantusiidae: Xantusia vigilis (RAG1: AY662642; C-mos: AF148703), Lepidophyma sylvatica (α-enolase: AY218079), Amphisbaenidae: Amphisbaena cubana (C-mos: AY487346), Amphisbaena xera (RAG1: AY662619), Rhineuridae: Rhineura floridana (RAG1: AY662618; C-mos: AY487347), Bipedidae: Bipes biporus (RAG1: AY662616; C-mos: AF039482), Trogonophidae: Trogonophis wiegmanni (RAG1: AY662617; C-mos: AY444025), Lacertidae: Eremias sp. (RAG1: AY662615), Lacerta kulzeri (C-mos: AF148712), Mesalina guttulata (α-enolase: AY218056), Teiioidea: Aspidoscelis tigris (RAG1: AY662620), Kentropyx calcarata (C-mos: AF420864), Tupinambis quadrilineatus (α-enolase: AY218076), Gekkonidae: Gekkoninae: Gekko gecko (RAG1: AY662625; C-mos: AY444028), Gehyra mutilata (α-enolase: AY218045); Pygopodinae: Lialis burtonis (C-mos: AF090850), Lialis jicari (RAG1: AY662628), Cordylidae: Cordylus polyzonus (RAG1: AY662643), Cordylus cordylus (C-mos: AF148711), Scincidae: Eumeces inexpectatus (RAG1: AY662632; HOXA13: AF083100; α-enolase: AY218075), Eumeces skiltonianus (C-mos: AF315396), Helodermatidae: Heloderma suspectum (RAG1: AY662606; C-mos: AY487348), Dibamidae: Dibamus novaeguinea (C-mos: AY487349), Dibamus sp. (RAG1: AY662645), Anguidae: Anniella pulchra (RAG1: AY662605; C-mos: AY487350), Varanidae: Varanus dumerilii (HOXA13: AF083102), Varanus rosenbergi (C-mos: AY099976), Varanus griseus (RAG1: AY662608), Serpentes: Pythonidae: Liasis savuensis (C-mos: AF544726), Cylindrophiidae: Cylindrophis ruffus (RAG1: AY662613), Iguanidae: Leiocephalus carinatus (RAG1: AY662598), Sauromalus ater (RAG1: AY662591).


