1 Introduction
The corallivorous sea star Acanthaster planci (Linnaeus, 1758), so-called crown-of-thorns starfish (COTS), is often seen as a major threat for coral reefs [1]. This species has an unstable population dynamics. The density of individuals can switch from a ‘normal’ situation with tens of individuals per square kilometer to an outbreak of thousands of COTS arriving on the reefs and devastating the coral community. The mechanisms of outbreaks are still poorly known. Sometimes the outbreaks seem to arise from several cohorts after multiple recruitment events [2]. However, along the Great Barrier Reef (GBR) in Australia, ‘primary’ outbreaks are the origin of successive ‘secondary’ outbreaks. The mechanisms of outbreaks seem to be a response to an increase in terrestrial nutrients discharges (that may be natural or enhanced by human activities), which triggers phytoplankton blooms, thus favouring the survival of A. planci larvae and consequently the recruitment of juveniles [3]. The decrease of natural predators such as the Giant Triton (Charonia tritonis), the Bullmouth Helmet (Cypraecassis rufa) or the Giant Helmet (Cassis cornuta) (Gastropoda, Mollusca), which are over-harvested by collectors, also favours A. planci adults survival in low density.
Acanthaster planci is distributed all over the tropical areas from the East Pacific Ocean to the West Indian Ocean. Individuals from the North Indian Ocean are blue rather than red-green to red-brown, as observed in the Pacific [4]. Outbreaks also occur in the West Indian Ocean and the Red Sea. In Mayotte (Comoros), where outbreaks have been documented (B.T., pers. obs.), the authorities are seriously worried about the coral reefs health, since healthy reefs provide essential tourist appeal. Given that the detection of environmental factors listed above is difficult to implement [2], the best way to manage A. planci populations during outbreaks is to collect sea stars by hand. However, if some larvae transported by ocean currents from a distant source regularly supply the population, then collecting is only a temporary solution to outbreaking events. The larval stage usually lasts several weeks, but it can be extended when survival conditions are especially unfavourable. That suggests strong dispersal ability and high gene flow between populations connected by ocean currents.
To date all population genetics studies on A. planci were carried out from allozyme variations in the Indo-West Pacific and the Pacific Ocean. They revealed a low differentiation level between populations from highly connected reefs and across the Pacific Ocean [4–6] and a strong genetic connectivity between Japan and the GBR [5,7]. No significant genetic differentiation was observed between outbreaking populations occurring 10 or 15 years apart [7,8]. However, genetic differentiation was observed among western and eastern Australian populations and among several small reefs isolated by local ocean currents [5,9]. The genetic differentiation between Indian and Pacific Oceans is relatively strong, since the genetic distance between A. planci populations from the two oceans was greater than that between A. planci from the Pacific Ocean and its sibling species A. brevispinus Fisher, 1917, a short-spined form living on soft bottoms between Indo-Pacific reefs [10]. Genetic data already confirmed the distinction between these two species. Despite their interfertility, hybrids have a low viability and developmental abnormalities [11,12]. Acanthaster planci seems to have evolved from an A. brevispinus-like ancestor during the Pleistocene [13]. The divergences observed with allozyme data appear therefore incongruent with the systematics of the genus Acanthaster, and this fact was neither confirmed nor checked by the study of different molecular markers.
There is therefore a need for more accurate genetic tools for studying populations of A. planci. DNA markers afford a wider panel of variability level than allozymes do and may be less prone to selection. Mitochondrial DNA (mtDNA) in particular is commonly used for phylogeographic study [14], since several universal primers are published and specific ones exist for asteroids [15,16]. Its complementarity to allozyme markers for phylogeographic inference mainly relies on the possibility of using evolutionary relationships among alleles after sequencing.
The aim of the present paper is to provide an assessment of the variability and the accuracy of the phylogeographic signal of three mitochondrial loci. A first insight into a mitochondrial phylogeography of the genus Acanthaster and an assessment of the divergence within the West Indian Ocean A. planci populations are also presented.
2 Material and methods
Samples of A. planci from Madagascar, Kenya, and Mayotte were collected between 2002 and 2005 (Fig. 1) from 10 to 2 m of depth. Gonads were collected and stored in 95% ethanol. Genomic DNA was extracted using Aquapure© Genomic DNA Isolation Kit (Bio-Rad) following the manufacturer's protocols. All mixtures were stored at −20 °C and used as a template for polymerase chain reaction (PCR). A 566-bp fragment of the 16S rDNA gene (16S) was amplified with forward primer 16SF2 (5′GTTTCGCCTGTTTACCAAAAACAT 3′) and reverse primer 16SR2 (5′AGGTTTTTCTGATCCAACATCG3′). A fragment coding for five tRNAs (Ala, Leu, Asn, Gln, Pro) (450 bp) and a part of the Cytochrome Oxydase subunit I (COI) (637 bp) were amplified as described in Knott and Wray [17]. PCR reactions were performed in 20 μl of a solution containing 25 ng of total extracted DNA, 4 nmol of dNTPs, 20 pmol of each primer and 0.5 U of FlexiGoTaq© DNA polymerase in the 1 X PCR buffer (Promega). The cycling parameters were 7 min at 95 °C, 35 cycles of 95 °C for 40 s, 30 s at 55 °C for the 16S locus and 45 s at 46 °C for the two other loci, followed by 2 min at 72 °C and a final extension of 7 min at 72 °C. PCR products were sequenced by industry and sequences were submitted to EMBL database under accession numbers AM410947 to AM410962. The level of polymorphism was compared among loci using the same set of seven individuals from the West Indian Ocean successfully sequenced at the three loci. To this subset of individuals, we added the sequences of the two complete mitochondrial genomes of A. planci and A. brevispinus (accession numbers AB231475 and AB231476). The DNA polymorphism parameters were estimated with DnaSP 4.10.8 [18]. Phylogenetic relationships between all sequenced individuals were estimated with the maximum-parsimony method (MP), a distance method using the neighbour-joining algorithm (NJ) and the maximum-likelihood method (ML). The best models of nucleotide substitution were determined with the program MrAIC [19] and turned to be Tamura–Nei (TN93) [20] for the 16S locus and Hasagawa–Kishino–Yano (HKY) [21] for COI and tRNAs loci. The MP and NJ trees were built with the program MEGA version 3.1 [22]. Given that the HKY model is not implemented in MEGA, all NJ trees were calculated with the TN93 distance. ML trees were built using PHYML [23] with the models of nucleotide substitution determined by MrAIC for each locus. Bootstrap values were calculated from 1000 replicates. Acanthaster planci sequences from Japan (AB231475) and Mexico (AF217324) as well as A. brevispinus sequence (AB231476) were added to the analysis. Based on their genetic proximity to Acanthaster, sequences of Meridiastra fissura (AY370740) were used to root the 16S gene tree and some of Patiriella calcar (U50046) for COI and tRNAs. Average pairwise distances between clades were estimated and rate constancy was checked with the Tajima's test of relative rate [24] implemented in MEGA. Minimum-spanning networks (MSN) of haplotypes were built using TCS [25].
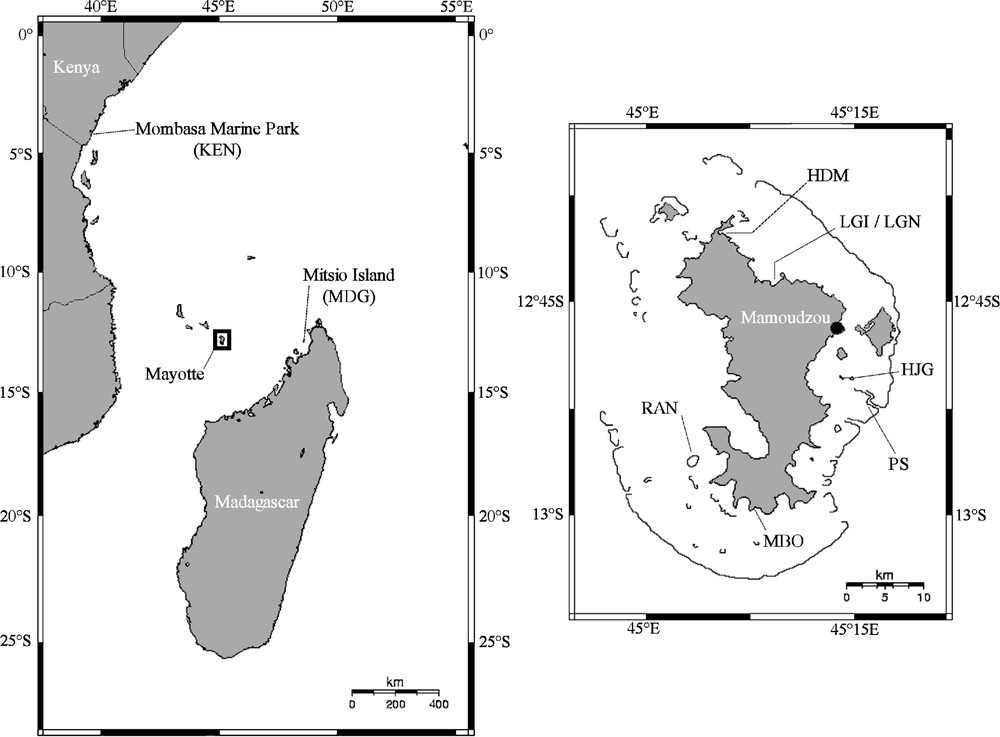
Geographical locations of A. planci samples. Left: samples from the north of Mozambique Channel: KEN, Mombasa Marine National Park, Kenya; MDG, Marie reef on Mitsio Island, Madagascar. Right: samples of A. planci from Mayotte island: HDM, Handréma Bay; LGI & LGN, Longoni Bay; HJG, Hajangua reef; PS, ‘Passe en S’; MBO, M'Bouini Bay; RAN, Rani reef.
3 Results
3.1 Overall mtDNA polymorphism
Considering the reduced set of individuals sequenced at the three loci, mtDNA sequencing revealed different levels of polymorphism depending on the region (Table 1A). Within the genus Acanthaster, the 16S and COI loci contain about 14% of segregating sites (five haplotypes), whereas tRNA only has 5% (three haplotypes). Haplotype and nucleotide diversities are of the same order of magnitude for 16S and COI (, and ) and were lower for tRNAs (, and ). At all three loci, more than 70% of segregating sites were singletons (mutations observed on only one sequence).
(A) DNA polymorphism of the genus Acanthaster within the reduced set of 9 individuals sequenced at all loci: 16S, tRNAs and COI. N: number of sequences; S: percentage of segregating sites; η: percentage of singletons; π: nucleotide diversity; n: number of haplotypes; : haplotype diversity. (B) Average divergences among clades and within West Indian Ocean (WIO) clade using the Tamura–Nei distance [20] on the complete sequence set. PO: Pacific Ocean
| A. | 16S | tRNAs | COI |
| N | 9 | 9 | 9 |
| S (%) | 14 | 5 | 15 |
| η (%) | 70 | 74 | 79 |
| π | 0.039 | 0.014 | 0.040 |
| n | 5 | 3 | 5 |
| 0.722 | 0.417 | 0.722 | |
| B. Div (%) | 16S | tRNAs | COI |
| A. planci / A. brevispinus | 11.3 | 4 | 12.1 |
| PO/WIO | 9 | 3 | 7.8 |
| within WIO clade | 0.2 | 0 | 0.3 |
3.2 Phylogenetic relationships and average divergences
The same tree topology was obtained using the three methods (NJ, MP and ML) and considering all loci. All tree branches are supported by high bootstrap values (Fig. 2). The genus Acanthaster is monophyletic and splits into an A. planci clade and A. brevispinus. The A. planci clade splits into a West Indian Ocean cluster and a Japanese cluster. The Mexican sequence available for tRNA only is closely related to the Japanese one, with which it forms a ‘Pacific Ocean’ clade distinct from the ‘West Indian Ocean’ one, which needs to be confirmed by additional sequences of the two other loci. The 16S and COI trees display similar average divergences (11.3% and 12.1%, respectively) between A. planci and A. brevispinus clades (Table 1B), whereas tRNAs shows a lower divergence (4%). The Tajima's relative rate test did not reject the molecular clock hypothesis () allowing us to roughly estimate the elapsed time since the divergence of the different clades. Applying the substitution rates established for Panamanian oreasterid species (5% divergence per million years (Myr) for the COI and 2.2% per Myr for tRNAs [15]) and assuming that theses rates are valid for Acanthaster (supported by the clustering of Oreaster with Acanthaster [26]), the average divergences between the sibling species, 12.1% (COI) and 4% (tRNAs) respectively correspond to 2.4 Myr and 1.8 Myr. Similarly, the divergence times between the two subclades of A. planci are estimated at 1.56 Myr and 1.36 Myr for COI and tRNAs, respectively. The variability observed within the West Indian Ocean clade was weak (or null) (Table 1B); however COI expressed the largest one. All haplotypes from Kenya were shared with Mayotte (Fig. 2B).
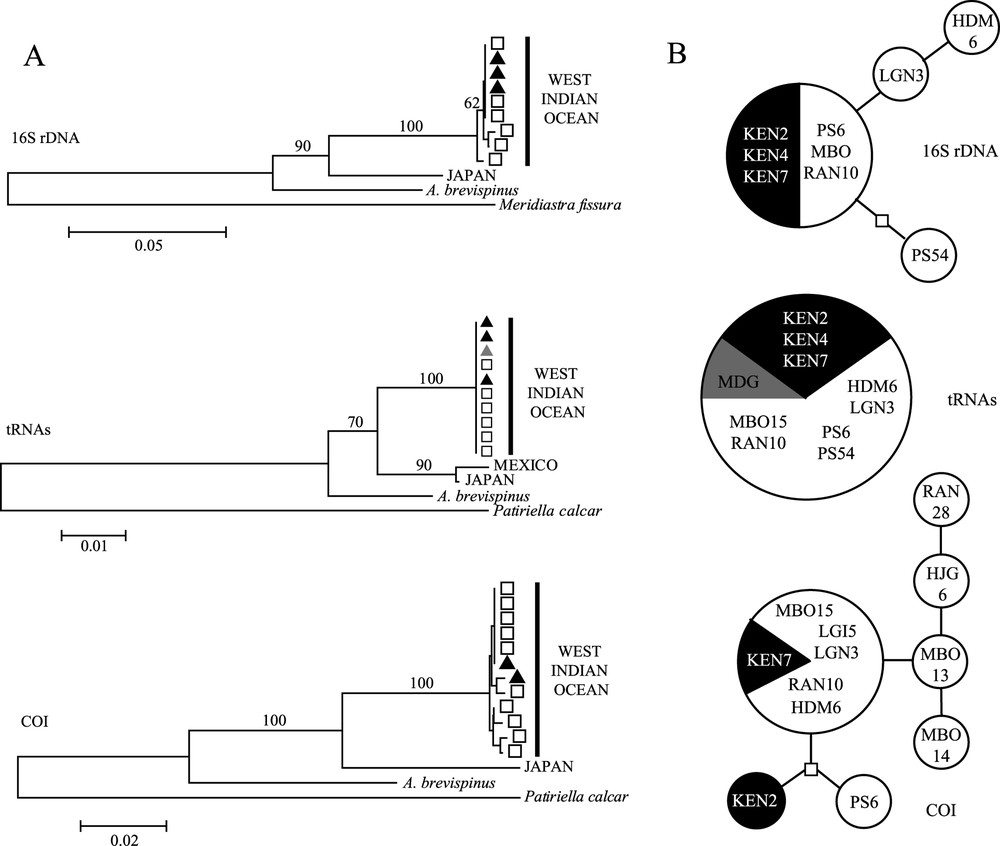
(A) Neighbour-joining trees of each locus, with bootstrap values indicated on the branches: black triangles are for Kenyan haplotypes, grey triangle for Madagascar, white squares for Mayotte. (B) The relationships within the West Indian Ocean clade of each locus are represented by a minimum spanning network of haplotypes: the circle size is proportional to the number of individuals displaying the haplotype (1, 6, or 10 individuals). Black slice/circles are haplotypes from Kenya; grey slice from Madagascar; white slices/circles are haplotypes from Mayotte.
4 Discussion
4.1 Evolutionary history of the genus Acanthaster
Our first phylogeographic data confirmed that Acanthaster forms a clade split into two subclades, A. planci and A. brevispinus, in agreement with Yasuda et al. [26], who analysed a single sequence from each species together with several published asteroid COI sequences. However, this contradicted allozyme data [10] that provided shorter genetic distances between A. planci and its sibling species in the Pacific. Allozymes are preferably used for intraspecific studies at the population level. The lack of allozyme differentiation between reproductively isolated species may originate from convergent phenotypes. In comparison, mtDNA provides a much stronger phylogenetic signal.
Within A. planci, our mitochondrial data confirmed the morphological and allozyme differentiation observed between Indian and Pacific populations. The estimations of the divergence time between these two subclades using both markers are congruent (1.56 Myr (COI) and 1.36 Myr (tRNAs)) and consistent with the time estimated by Benzie [10] (1.6 Myr). Indian Ocean populations may actually be genetically isolated from the Pacific ones. Within the West Indian Ocean clade, all three locations share identical or closely related haplotypes as indicates the low within-clade mean divergences (0.2% (16S) and 0.3% (COI)).
4.2 Accuracy of mtDNA in studying A. planci phylogeography
All tree topologies from mtDNA are congruent with systematics, morphology (colour morphs) or biogeographical patterns previously published for Acanthaster. Considering the set of individuals sequenced at the three loci, tRNAs evolve three times more slowly than COI and 16S. The latter two markers seem to display similar levels of polymorphism, suggesting that they evolve at the same rate within the genus Acanthaster. That feature contrasts with the trends observed for a majority of species, since COI usually evolves faster [27]. The assessment of extant polymorphism may be biased by the reduced size of this subset of samples, since actually COI appears as the most polymorphic locus when the whole COI sequence set is considered (Fig. 2). Alignment of the protein coding COI sequences from distant species is much easier than alignment of 16S sequences, which contain insertions and deletions. These two features usually make COI a better mitochondrial marker than tRNAs and 16S for phylogeographic inferences. However, in the present study, all three loci appeared as good candidates for worldwide study of Acanthaster phylogeography and good complementary tools to more variable and independent loci usually required for population genetics study at small geographic scales such as nuclear microsatellites [28] or introns [29,30].
Acknowledgements
UMR DIMAR financially supported this work. We are very grateful to the Service des pêches de Mayotte for logistic and financial support through the GIS LAGMAY, and to E. Turak and S. Visram for collecting samples in Madagascar and Kenya, respectively. Thanks are also due to E.F. Dick who kindly helped to improve the English text. We finally thank the anonymous reviewers for helpful comments on the manuscript.


