1 Introduction
Quinol-oxidizing complexes – the cytochrome bc1 complex of mitochondria and bacteria or its chloroplast counterpart, the cytochrome
While numerous data and mutation studies have provided a fairly good description of the Qo site from both cytochrome
Despite their similarities, bc1 and
The scarcity of mutagenesis data also originates from the fact that cyanobacteria do not tolerate severe alterations in the function of the
In C. reinhardtii the cytochrome
Here, we critically re-examine our previous attempts to obtain mutants of the Qi site, impossible to interpret in the absence of the 3-D structure. Mutant construction was based on sequence and structure homology between cytochromes
2 Results
2.1 Before the structure
In cytochrome bc1 the Qi site is formed by cytochrome b and most mutations altering the functionality of the Qi site target the amino-terminal part of cytochrome b or the de loop connecting helices D and E [4]. Cytochrome
Two residues in helix A from cytochrome
By site-directed mutagenesis of plasmid pWB encompassing the petB coding sequence [24], we substituted the conserved Gly37 of cytochrome
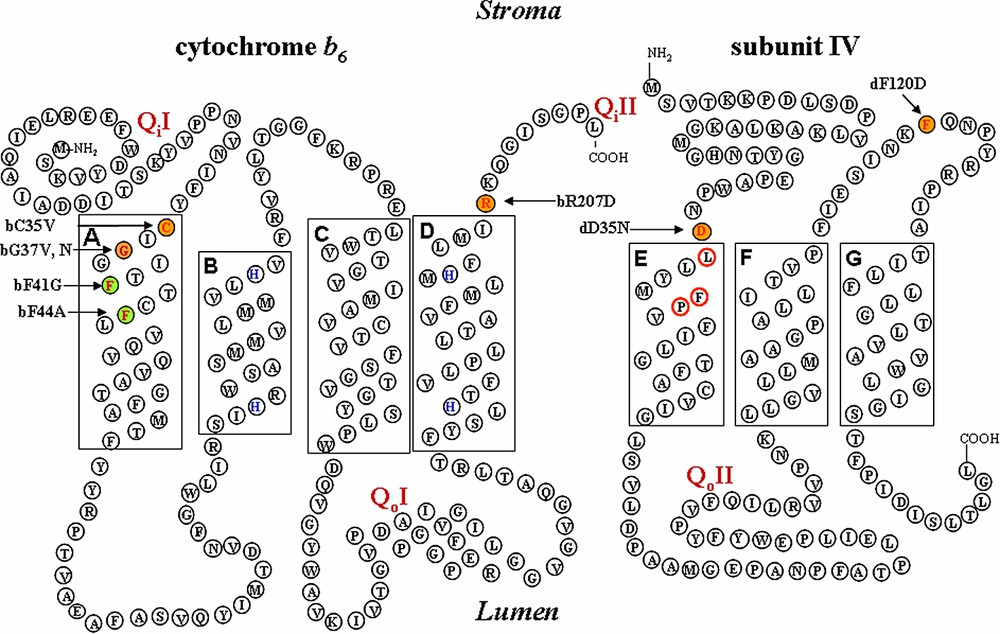
Schematic representation of cytochrome
These mutated petB genes were introduced to the chloroplast genome of C. reinhardtii by biolistic transformation, according to two strategies used in parallel (see Section 4). In the first one, deletion strains – here ΔpetB [24] – were used as recipient and transformed clones were selected on minimum medium for the recovery of photosynthetic capability, when the mutations preserve the function of the cytochrome
Two mutants (bF41G and bF44A) were recovered after transformation of strain ΔpetB, following the capability of the deleted strain to recover photosynthesis. Characterisation of these mutant strains did not reveal any drastic changes, since they accumulated the
By contrast, no phototrophic clones could be recovered upon transformation of the ΔpetB strain with petB genes carrying substitutions of the Gly37 residue, suggesting that these mutations are detrimental to photosynthesis. Consistently, spectinomycin-resistant transformants obtained from the wild-type recipient strain displayed fluorescence induction kinetics typical of mutants with impaired cytochrome
The structure of the cytochrome bc1 complex that became available in the late 1990s [9,11,30] (PDB accession 1bcc and 3bcc) allowed us to select other residues putatively contributing to the formation of the Qi site. We chose the residues Arg207 at the C-terminus of cytochrome
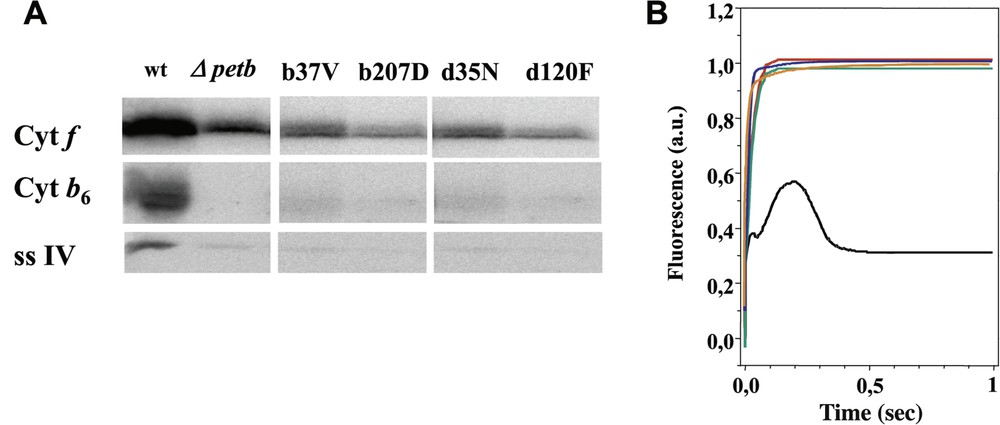
Panel A: Immunoblot against subunits
We also substituted a Val for Cys35, a residue specifically conserved in all cytochrome
2.2 After the structure
We have already pointed out how the unexpected discovery of the
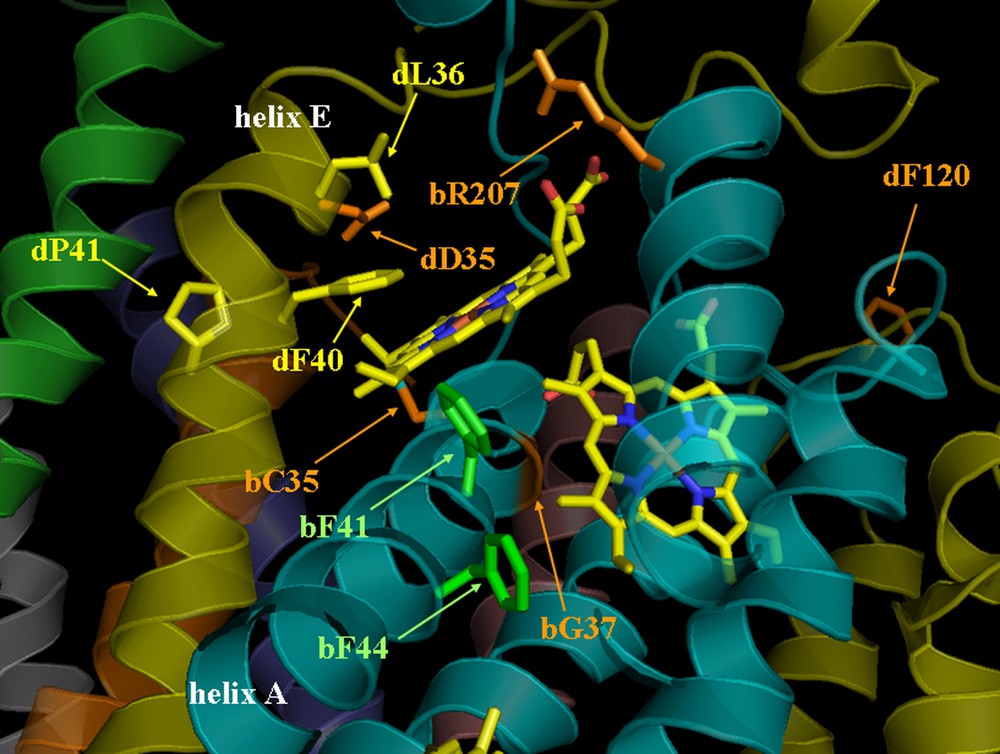
View of Qi site (1Q9 pdb data). Mutated residues are located in the structure; in orange: mutations leading to non-assembled complexes and non-photosynthetic strains (on cytochrome
View of Qi site (1Q9 pdb data). Mutated residues are located in the structure; in orange: mutations leading to non-assembled complexes and non-photosynthetic strains (on cytochrome
Our attempts to recover phototrophic clones upon transformation of the ΔpetD strain were unsuccessful, leading us to suppose that also these mutations were detrimental for photosynthesis. We then used the wild-type strain as a recipient for transformation experiments with plasmids carrying the specific mutations associated with the aadA cassette. Unexpectedly, the transformed clones recovered on antibiotic-supplemented medium retained wild-type fluorescence induction kinetics, even after they reached homoplasmy for the mutation (when all copies of the polyploid chloroplast genome carry the mutation while all wild-type versions of the petD gene have been eliminated). Paradoxically, the mutations therefore were not harmful to photosynthesis, even though the mutated genes were not able to rescue deletion mutants upon transformation.
3 Discussion
For more than ten years, we have tackled the ‘mystery’ of the Qi site. The resolution of the 3-D structure of the cytochrome
The mutated amino acids can now be located on the 3-D structure (Fig. 3) [8] in order to ascertain the structural effects of the mutations on the Qi site environment and to correlate it with the corresponding phenotype.
Gly37 of cytochrome
The detrimental effect of substitution for an Asp of residue Arg207 (Fig. 3, residue coloured in orange on the cytochrome
Substitutions of the two Phe residues of cytochrome
Two other mutations were carried out on subunit IV (Figs. 1 and 3), both of which yielded non-phototrophic mutants. Residue Asp35 (residue coloured in orange on the subunit IV, helix E in yellow) is in strong electrostatic interaction with residue Arg95 from the PetN subunit. Its substitution with Asn by removing the negative charge changes the local polarity, which seems important for the conformation of the site and the stability/assembly of the complex, as shown by the phenotype of the mutant. It is more difficult to explain the detrimental effect of the Phe120 → D mutation, since residue dPhe120 (coloured in orange on the subunit IV, fg loop in yellow) is located quite far away from the Qi site and does not seem to interact with other cofactors. However, Phe120 is close to a positively charged patch (Arg112 on cytochrome
Our successful attempts to obtain Qi site mutants were based on the available 3-D structure. However, we could not rescue the ΔpetD deletion strain for photosynthetic growth upon transformation with the mutated plasmids. Paradoxically, after selection for resistance to antibiotics, the dL36A, dF40H/Y/L and dP41A mutants nevertheless turned out to be capable of phototrophic growth. The molecular basis of this paradoxical observation is not fully understood yet and was first hypothesised to reflect the complex organization of the Qi site. As evoked above, the full structure of this site is particularly complex, since it requires interactions between two subunits, cyt.
4 Materials and methods
4.1 Strains, media, and growth conditions
A wild-type strain derived from strain 137C [35] and the deletion strains ΔpetD and ΔpetB [24] were used as recipient strains in chloroplast transformation experiments. WT and mutant strains were grown on Tris-acetate-phosphate (TAP) medium (pH 7.2) at 25 °C, under dim light (5–6 μE m−2 s−1), unless otherwise indicated.
4.2 Site directed-mutagenesis
Plasmids pdWQ and pdWB, encompassing the regions coding for subunit IV and cytochrome
Mutations Leu36Ala, Phe40Leu/His/Tyr and Pro41Ala were introduced in plasmid pWQ by PCR-mediated site-directed mutagenesis [36]. The aadA cassette conferring resistance to spectinomycin [25] was then inserted into the pWB or pWQ plasmids carrying the various mutations, respectively at the NsiI or EcoRV neutral restriction sites [2,26].
4.3 Chloroplast transformation in C. reinhardtii
Wild-type as well as ΔpetB and ΔpetD strains, respectively deleted for petB or petD genes, were transformed by tungsten particle bombardment according to Boynton et al. [37]. Transformants were selected either on minimum medium under dim light (60 μE m−2 s−1) or on TAP medium supplemented with spectinomycin (100 μg ml−1) under low light (10 μE m−2 s−1). They were submitted to several rounds of subcloning on selective medium until they reached homoplasmy, assessed by RFLP analysis of specific PCR amplification fragments.
4.4 Preparative and analytical techniques
For SDS-PAGE, membrane (or whole cells) proteins were resuspended in 100 mM dithiothreitol and 100 mM Na2CO3, and solubilised by 2% SDS at 100 °C for 1 min. Polypeptides were separated on a 12–18% polyacrylamide gel containing 8 M urea [38]. Immunoblotting was performed as described in Pierre et al. [15].
4.5 Fluorescence measurements
Fluorescence measurements were performed at room temperature on a home-built fluorimeter, using a light source at 590 nm. The fluorescence response was detected in the far-red region in the near IR region [26].
Acknowledgements
The authors are grateful to Yves Choquet and Jean-Luc Popot for their critical reading of the manuscript. Thanks are due to Daniel Picot for the fruitful discussion and the help in drawing Fig. 3. F.Z. whishes to thank Francis-André Wollman for his fundamental input in this study. A.L.L. was the recipient of a fellowship from the ‘Ministère de l'Enseignement supérieur et de la Recherche’, France.



Vous devez vous connecter pour continuer.
S'authentifier