1 Introduction
A few years ago, we published a special issue of the Comptes rendus de l'Académie des sciences [1] to celebrate the centenary of the independent discovery by Sergius Nawashin [2] and Léon Guignard [3] of the double fertilization process in plants. These pioneer authors independently observed for the first time the occurrence of a double fertilization in two different plant species, Turk's cap lily (Lilium martagon) and the perennial herb Fritillaria tenella. At that time no one was using model plants or model systems. Yet, this important discovery still corresponds to one of the key hallmarks in plant biology. Indeed, the double fertilization is unique to flowering plants among living organisms and permits the establishment of a new generation, from the zygote to the embryo included within the seed. The beginning of embryology research during the 20th century led to many novel aspects in developmental biology, notably in plants. Several books have been published on this topic [4], of which the monograph by Masheswari remains the historical reference in plant embryology [5]. In this paper, we summarize key features related to fertilization and the earliest developmental aspects of embryo tissues within the seeds of flowering plants.
2 Fertilization in angiosperms: a multi-step phenomenon
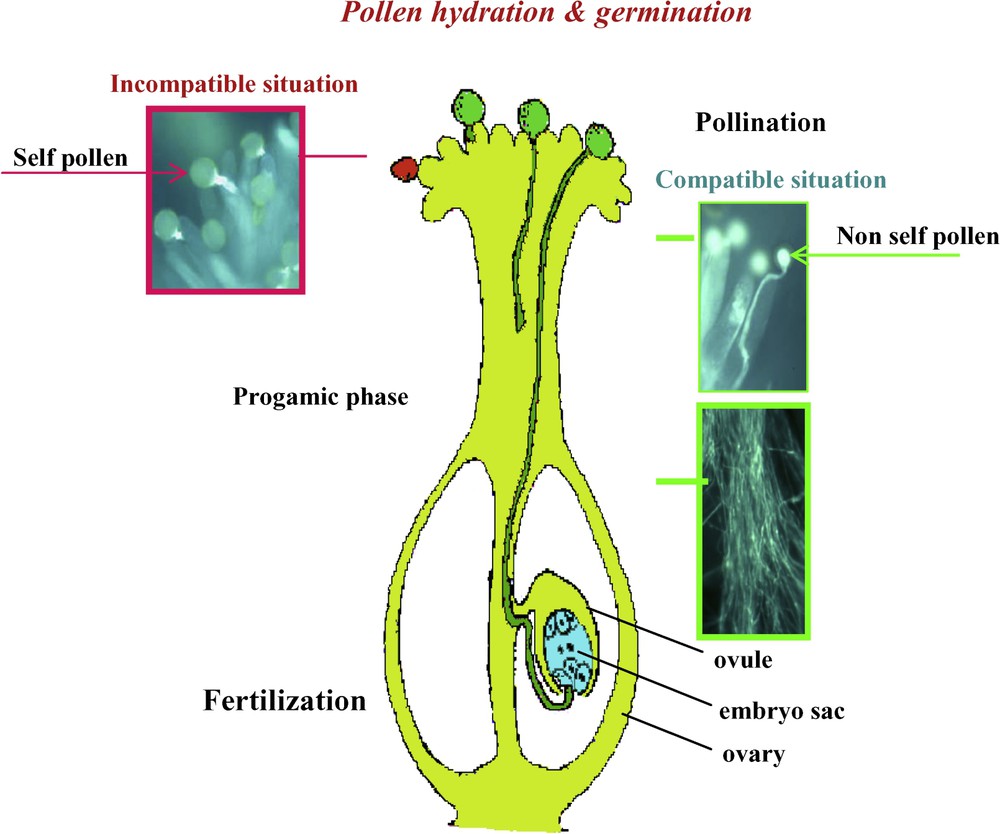
Fertilization in flowering plants (angiosperms) is a very complex process as compared with animal or algae systems. It consists of three successive phases (Fig. 1):
- – Pollination that corresponds to the transfer of a male nucleus containing unit, called male gametophyte or pollen grain, from the male organ, the anther, to the receptive female organ, the stigma surface of the pistil. Note that since plants are sessile organisms, sexual partners just meet by chance, sometimes with the aid of animals like insects (entomogamy), wind (anemogamy) and, more rarely, water (hydrogamy).
- – A progamic phase that includes all processes occurring from the landing of pollen grains on the receptive stigma to the time that the sperm cell reaches the egg cell. More precisely, this phase includes different processes such as adhesion of the pollen grain to pistil prior to water uptake, enzyme (e.g., cytochrome oxidase, cellulase, phosphorylase, ribonuclease, acid phosphatase, …) release and activation, and the preparation of pollen tube (PT) formation. In this tube, a second mitotic division often occurs leading to the formation of two sperm cells. Here, the PT acts as a sort of vehicle (sperm cells are not motile) carrying the two sperm cells to their target cells within the so-called “ovule” (a terminology that is somehow misleading in relation to animal or algae terminology). Ovules in angiosperms are maternal organs containing the female gametophyte, the embryo sac (a complex haploid and pluricellular structure containing two female gametes), the egg cell and the central cell (Fig. 1). During the course of evolution, about half of the angiosperm species have acquired specific recognition mechanisms, which strongly limit or even prevent self-fertilization. The most sophisticated and widespread of these mechanisms is self-incompatibility (SI), a process leading to the rejection of self-pollen by the pistil. SI is controlled by a single multiallelic locus, the S-locus (S standing for self-incompatibility) and several plant systems have been carefully analyzed for this aspect, e.g., Brassica or Papaver [7–9].
- – The third phase, referred to as syngamy, is the last and decisive phase of fertilization, which corresponds to the unique double nuclear fusion event in angiosperms. Here, the first sperm nucleus fuses with the egg nucleus (at the origin of the zygotic embryo), while the second sperm nucleus fuses with the two polar nuclei of the central cell of the embryo to form the triploid endosperm (Fig. 2). These three steps, and especially the syngamy step, are now well described and largely understood both at the cytological and molecular levels since the original description by Nawashin and Guignard [6,7]. Lilium martagon and Fritillaria tenella were originally used to discover and first describe the double fertilization process using the classical microscope [2,3]. Later, a new source of excitement came with the advent of electron microscopy and its use to elucidate the syngamy, notably with the aid of cotton (Gossypium hirsutum) as a model. These crucial observations made by the group of W. Jensen first showed that the two male gametes are real cells devoid of cell walls [10]. Second, they documented that the plasma membrane of the gamete cells is in close contact with the inner plasma membrane of the vegetative cell that surrounds them in the pollen grain. In addition, the nucleus of the vegetative cell seems to be physically associated to this structure, forming the so-called ‘male germ unit’ [11]. In more recent studies, maize (Zea mays) was used to isolate both male and female gametes (sperm, egg and central cells) [12] to establish an in vitro system mimicking in vivo conditions [13]. Caryogamy was clearly examined by using a combination between an in vitro assay and a three-dimensional image reconstruction from electron microscopy data [14]. The first detectable cellular event taking place after gamete fusion was an increase in the concentration of cytosolic Ca2+, as occurs in animal gamete fusion [15]. This rise occurred after the establishment of gamete cytoplasm continuity. With the aid of an extracellular vibrating probe, this allowed one to demonstrate that a calcium influx is triggered and propagates in the zygote as a wave front [16].
The steps of fertilization in flowering plants.
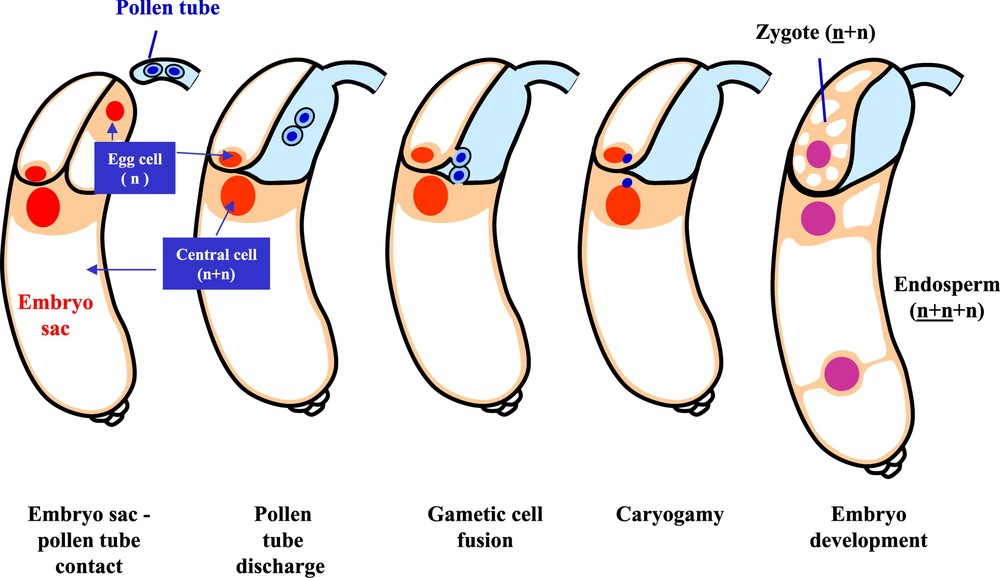
Events of double fertilization. The interaction between the male (pollen tube) and female (embryo sac) gametophyte is followed by gametic cell fusion, caryogamy and embryonic development.
3 Molecular and cellular biology of fertilization
The question arises as to whether Ca2+ accumulation is necessary and/or sufficient to trigger egg activation and the initiation of development. The sole use of in vitro approaches could not help answering this question. Therefore, several complementary approaches have been developed including (a) molecular analyses of cDNA libraries prepared from isolated gametes [17] and zygotes [18], (b) new developments including life imaging using Arabidopsis (Arabidopsis thaliana) as a model system [19,20], and (c) the search for mutants affected in fertilization. This allowed a novel class of MYB factors controlling sperm-cell formation to be identified [21]. Furthermore, the female gamete was found to be able to regulate the delivery of male gametes. In fact, in the feronia and sirene Arabidopsis mutants, embryo sac development is not affected and synergid differentiation appears to be normal. However, after pollen tube penetration in the embryo sac a failure in sperm discharge is evidenced [22]. After the recent characterization of FER/SIR (FERONIA/SIRENE) as a synergid-expressed, plasma-membrane-localized receptor kinase, a putative model for PT reception has emerged [23]. In this model, when the pollen tube reaches the synergids, a ligand issued from the PT binds to the FER/SIR extracellular domain, triggering a signaling cascade that enables the female gametophyte to prepare itself for fertilization. It is noted that in the gametophytic factor2 mutant the synergids do not degenerate while they normally do so during the fertilization process. The corresponding gene has been cloned and shown to encode a DNAJ chaperonin localized to the mitochondria. Thus, most presumably synergid cell death depends on the involvement of some mitochondrial function [24]. Experimental ablation studies in the plant Wishbone flowers (Torenia fournieri) suggest that only intact synergids can attract the PT to the ovule, underscoring the importance of both synergids in PT receipt. Here, the persistent synergid is important for attraction and the other typically degenerated receptive synergid is important for entry of the PT in the embryo sac [25]. A critical role for actin coronas has also been observed in degenerated synergids, both near the egg nucleus and ending near the central cell [26]. Also, a MYB protein has been identified as a transcriptional regulator of genes expressed in synergid cells and required for the formation of the filiform apparatus and PT guidance [27]. Furthermore, the highly expressed Generative Cell Specific1 (GCS1) protein located in the sperm cell membrane proved indispensable for the fusion of the two sperm cells with the egg cell and the central cell [28]. Only fertilized eggs proved to be associated with actin coronas, in marked contrast to unfertilized supernumerary egg cells lacking an actin corona as occur in the maize mutant indeterminate gametophyte1 (ig1) [29].
Nuclear migration within the central cell generally precedes fertilization. Furthermore, egg cell fertilization precedes central cell fertilization both in Arabidopsis and maize, two well studied plant models [19,30]. Central cell guidance (ccg), a new mutant defective in micropylar PT guidance has been identified [31]. The CCG gene encodes a nuclear protein with an N-terminal conserved zinc β-ribbon domain that is functionally interchangeable with that of TFIIB (a basal transcription factor) in yeast, suggesting that CCG might act as a transcription regulator for PT guidance.
To date there are no reports pointing toward a sperm cell dimorphism in Arabidopsis. In fact, this exceptional phenomenon suggesting a preferential fertilization has only been reported in Ceylon Lead wort (Plumbago zeylanica) and in B chromosome line of maize [32,33]. However, new experimental data support the occurrence of preferential fertilization as a general phenomenon [34].
4 Early embryo and endosperm development
The developmental events leading from the two single cells, the zygote and the fertilized central cell, to two multi-cellular, highly differentiated organs with elaborate body plans, namely the embryo and the endosperm, are generally referred to as early development. This development provides the structures necessary for the accumulation of reserve storage molecules, which in the case of the embryo are used for further development after germination.
The development of plant embryos is fundamentally different from that in animal systems. Firstly, plant embryogenesis is not a distinct process leading to the formation of a miniature version of the adult organism containing at least primordia of all its future organs. It is rather the beginning of a continuous developmental process interrupted temporarily by drying and dormancy [35]. Secondly, the elaboration of the body plan is not based on cell lineage but on the position of individual cells within the embryo [36], gradients of hormones or other signaling molecules determining the fate of individual cells [37]. Thirdly, plants have a unique mode of cytokinesis involving plant-specific structures such as the phragmoplast or the preprophase band [38]. Finally, the cells of the plant embryo are not mobile and show neither the long-distance homing nor the interstitial migration seen in animal and in particular in vertebrate embryos [39].
In both monocots (e.g., maize) and dicots (e.g., Arabidopsis) early embryo development is marked by three major events corresponding respectively to the acquisition of an apico-basal polarity, the differentiation of an epidermis and the formation of the shoot and root meristem [40]. The origin of apico-basal polarity is a matter of debate, the question being whether this polarity is established de novo in the embryo or inherited from the egg cell. Polarity is clearly established after the first cell division of the embryo due to clear cytological differences between a cell rich in cytoplasm, which gives rise to the embryo proper and a strongly vacuolized cell at the origin of the suspensor [41]. Marker genes specifically expressed in either cell, such as WOX2 in the upper cell and WOX8 in the lower cell, support the polarization of the two-celled embryo [42]. While some authors claim that the plane of the first division, which is perpendicular to the embryo axis, is at the origin of embryo polarity [40], others argue that exceptions with longitudinal or oblique first divisions exist implying that polarity is already established in the zygote or egg cell [35]. In this context it is interesting to note that a rather dramatic shift in cell polarity takes place in the maize egg cell upon fertilization. While the nucleus and most of the cytoplasm are located at the micropylar half of the egg cell prior to fertilization, they are found in the antipodal half afterwards, matching the polarity of Arabidopsis or the plant shepherd's purse (Capsella bursa-pastoris L.) [43,44]. This observation argues in favor of an influence of the fertilization process itself on the polarity of the zygote. The remaining events of early embryo development are discussed elsewhere in this issue [45].
The development of the endosperm starts with the fertilized central cell, which can undergo a cellular, nuclear or mixed (helobial), type of development. Both the Brassicaceae (Arabidopsis) and the Gramineae (barley, maize) have an endosperm of the nuclear type, even though it is believed that this endosperm arose independently during evolution in these two lineages [46]. In nuclear endosperm, the initial endosperm nucleus divides repeatedly without cell wall formation, resulting in a coenocytic endosperm, in which the nuclei are distributed in the periphery of a giant single cell, surrounding a central vacuole (Fig. 3). While this stage is also referred to as syncytial endosperm, we wish to avoid this term, because it implies for certain readers that the multinucleate cytoplasm arose from cell fusion rather than from nuclear division. The nuclear divisions are highly synchronized resulting in Arabidopsis in over 200 nuclei after eight rounds of division. While the first three rounds of division are fully synchronized, a slight delay of some divisions during the following rounds allows the definition of three mitotic domains. While nuclei are still dividing synchronously within the micropylar endosperm (MCE) surrounding the embryo and within the peripheral endosperm (PEN) in the central chamber, no more divisions are observed in the chalazal endosperm (CZE) [47]. Specific marker lines with domain-specific expression confirm the regionalization of the endosperm prior to cellularization [48,49].
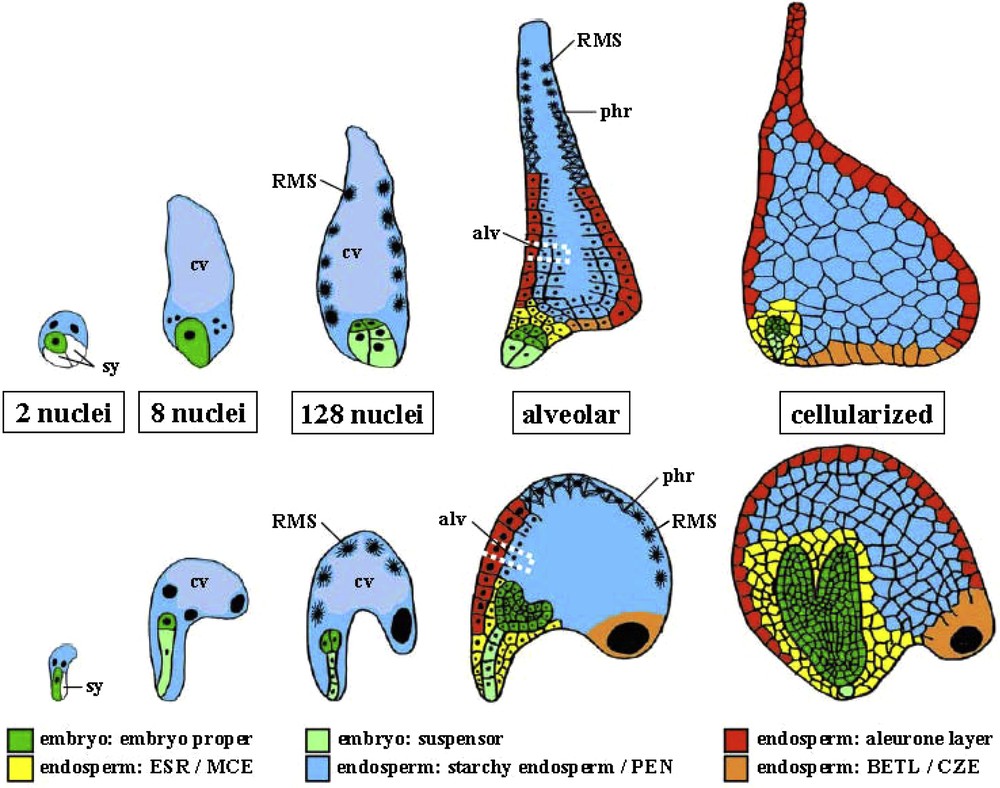
Early embryo and endosperm development in maize and Arabidopsis. Schematic drawings of embryo and endosperm in maize (top) and Arabidopsis (bottom) at 5 stages: 2, 8 and 128 nuclei in the endosperm, alveolar and cellularized. Note that not all nuclei are in the plane presented by the drawing. The central vacuole (cv) of the single celled endosperm coenocyte is indicated in light blue at two stages. The radial microtubule system (RMS) is indicated around free nuclei. One of the alveoli (alv) is surrounded by a dotted white line. Synergids (sy) are not colored. BETL, basal endosperm transfer layer; CZE, chalazal zone endosperm; ESR, embryo surrounding region; MCE: micropylar cellularized endosperm; PEN, peripheral or central endosperm; phr, phragmoplast.
The cellularization process starts at the final round of coenocytic mitosis as a wave in the MCE, progressing through the PEN and CZE at different rates and with significant variations between the domains [50]. The first step of cellularization is the formation of a radial microtubule system (RMS) emanating from the surface of endosperm nuclei and defining nuclear cytoplasmic domains (NCD). The second one is the formation of “free growing” phragmoplasts at the zones where growing NCDs contact each other. The third one is alveolar cell wall formation leading to a tube around each nucleus with its open end toward the central vacuole. The last one is the formation of a periclinal cell wall at the next nuclear mitosis [50]. Several genes have been shown to be involved in the cellularization process, including members of the FIS polycomb group [51] and the MADS box transcription factors AGL62 [52]. In maize, barley (Hordeum vulgare) and rice (Oryza sativa) the early steps of endosperm development and in particular the formation of the coenocyte and the mechanisms of early cellularization are very similar to Arabidopsis [53,54], despite major morphological differences between the embryo surrounding region [55,56], the basal endosperm transfer layer [57,58], the starchy endosperm [59] and their functional equivalents MCE, PEN, CZE in Arabidopsis (Fig. 3).
After cellularization the Arabidopsis endosperm enters directly into endoreduplication, while numerous additional cell divisions occur in cereals prior to endoreduplication [60]. Following endoreduplication the endosperm is consumed rapidly by the developing embryo, while the cereal endosperm starts to accumulate starch and protein reserves, which will nourish the embryo after germination, as discussed in this issue [61].
The endosperm and embryo develop in parallel to form the seed, but little is known about the coordination between these two organisms as well as the coordination with the surrounding seed coat. Signaling from the embryo to the endosperm was suggested by the characterization of the Arabidopsis cdc2A (also called cdka;1) mutant, which shows a strong paternal effect. In mutant pollen, only one sperm cell, instead of two, is produced. The pollen is viable but can fertilize only the egg cell and not the central cell. However, the unfertilized endosperm develops, suggesting that a previously unforeseen signal from the fertilized egg initiates proliferation of the central cell [62]. Evidence for communication between the endosperm and the seed coat comes from the analysis of the genes Haiku2 (Iku2) and Miniseed3 (Mini3) encoding a leucine-rich-repeat kinase and a transcription factor of the WRKY family, respectively [63]. Both mutants exhibit decreased endosperm size, along with a decrease in cell elongation in the seed integuments [64].
5 Balance of paternal and maternal genome contribution to embryo and endosperm
Seed development is strongly influenced by the balance of the paternal and maternal genomes. In wild type seeds the diploid embryo generally contains one maternal and one paternal genome (1m:1p). In contrast, the triploid endosperm has an inbuilt imbalance of two maternal to one paternal genome (2m:1p). Deviation from these ratios, for example in interploidy crosses between diploid and tetraploid lines, frequently leads to defects or abortion of the developing seeds. Generally the endosperm seems more affected than the embryo by developmental aberrations [5]. While beneficial effects of interploidy crosses on seed development have been documented in certain species, they remain an exception [65]. The first conclusive evidence for the importance of the balance of maternal to paternal genomes for normal seed development came from interploidy crosses involving the indeterminate gametophyte (ig) mutation in maize, which allowed to rule out a role of total ploidy levels or of the genome ratios between embryo, endosperm and the seed coat [66]. Depending on the direction of the interploidy cross, the progeny can exhibit either maternal genome excess (MGE, embryo: 2m:1p, endosperm: 4m:1p) or paternal genome excess (PGE, embryo: 1m:2p, endosperm: 2m:2p). Phenotypic analyses of Arabidopsis seeds obtained by reciprocal crosses between diploid and tetraploid as well as between diploid and hexaploid lines showed that MGE and PGE produce complementary phenotypes. Thus, MGE inhibits endosperm development by premature cellularization, causing in turn a developmental delay or arrest of the embryo. In contrast, PGE promotes growth of the endosperm by accelerated mitoses and delayed cellularization, leading to larger embryo sizes [67]. Similar results were recently obtained in maize. It appears that MGE seeds are very small and generally abort, their endosperms cellularize earlier, they enter earlier into endoreduplication and they accumulate significant amounts of starch. In contrast, PGE seeds are slightly bigger than MGE seeds though they are still smaller than wild type seeds, undergo an extended period of cell proliferation, show little endoreduplication and accumulate little starch [68,69]. The molecular mechanisms causing the temporal deregulation of the ontogenic program of endosperm development and disturbing the balance between cell proliferation and cell differentiation are not known. However, there is agreement that parental imprinting may play an important role [51]. First evidence for this comes from methylation studies of the Fie1 (Fertilization independent endosperm1) and Fie2 genes in isolated gametes and fertilization products of maize showing that the exclusively maternal allelic expression during early endosperm development perfectly correlates with methylation of the 5′ end of the paternal alleles. There was no Fie2 methylation in sperm cells, suggesting that the methylation was actively established during or shortly after fertilization [70]. Histone modifications also seem to be involved since the analysis of endosperm with altered parental genome dosage indicated that the redistribution of the H3K9me1 heterochromatin marker from chromocenters toward euchromatin and interspersed heterochromatin concerned predominantly the maternal but not the paternal genome [71]. In addition, clustering of the HTR12 centromeric histone of paternal origin at one extremity of endosperm nuclei even after two divisions was interpreted as a segregation of the paternal chromatin from the maternal chromatin, which may prefigure parental genomic imprinting [72].
6 Development of unfertilized male and female gametes
Unfertilized gametes can develop to certain degrees and even give rise to haploid, generally sterile plants. Artificial doubling of chromosomes at the seedling stage prevents sterility and together with self pollination allows the production of perfectly homozygous plants, which is of a great interest for plant breeders. The development of the non-fertilized egg cell or gynogenesis naturally occurs in most Angiosperms. The frequency is generally very low, e.g. 0.1% in maize [73]. This frequency can be dramatically increased by the use of particular male parents (inducing lines) to reach for example 8.1% in maize [74]. While unfertilized sperm cells do not develop into embryos, immature microspores, the product of male meiosis, can be induced by various stresses to form haploid embryos in vitro, which can be regenerated into fertile plants after chromosome doubling. This process was originally termed androgenesis and more recently microspore embryogenesis. Its efficiency is strongly species and genotype dependent and can reach over 30% in rapeseed [75]. There are no reports on substantial development of unfertilized central cells other than in mutants.
The best characterized mutations allowing the development of unfertilized central cells concern members of the chromatin remodeling Polycomb group (PcG), which in wild type plants plays a major role in the arrest of female gametophyte development after fertilization [76,77]. In the seed, the complex is composed of the SET (Suppressor of variegation/Enhancer of zeste/Trithorax) domain protein MEDEA (MEA) [78], the VEFS domain protein FERTILIZATION INDEPENDENT SEED2 (FIS2) [79], the WD40 repeat proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE) [80] and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) [81]. Loss-of-function mutants of members of the complex show autonomous development of the central cell in the absence of fertilization. In addition, msi1 mutants also show a few divisions of the egg cell leading to early arrested non-viable haploid embryos [82]. The Polycomb Repressive Complex2 (PRC2) is thought to repress gene expression by methylation of histone H3 at K27 (H3K27). Correlation of gene repression with the presence of the H3K27 marks has been demonstrated for three direct targets of the seed PRC2, namely Pheres1 (Phe1) coding for a MADS box transcription factor [83], Fusca3 (Fus3) coding for a B3 transcription domain factor [84] and Formin Homology Protein5 (FH5) encoding an actin nucleator involved in endosperm cellularization [85]. The regulation of the genes encoding members of the PRC2 is complex. Thus, MEA does not only auto-regulate its own transcription [86] but is also subject to paternal imprinting [87], just like certain other members of the PRC2 complex. On the other hand, the target gene Pheres1 is one of the rare genes known to be subject to maternal imprinting by the PRC2 [88]. Arabidopsis Glauce (Glc), whose molecular identity is not yet known, counterbalances the action of the PRC2 by promotion of fertilization-independent endosperm development and expression of paternally inherited alleles [89]. More recently, viable seeds consisting of a normal diploid embryo and an unfertilized endosperm have been obtained by fertilization of PRC2 mutants by hemizygous CDKA;1-YFP plants generating only one sperm cell. This result suggests a more general role of the PRC2 complex in balancing the contribution of the paternal genome in the triploid endosperm and has been used as an additional argument that the endosperm represents in evolution an extension of female gametophyte development rather than a supernumerary embryo [90].


