1 Lipid bodies in oleaginous plant/oil crops
In all cells, lipids required for energy supply are stored in lipid bodies (LB), also called lipid droplets or oil bodies. Oleaginous plants are cultivated essentially for oil production, for both nutritional and industrial use. In France, rapeseed (Brassica napus) and sunflower (Helianthus annuus) have been cultivated in 2007 over an area of . The most widespread sites for lipid accumulation in plants are seeds, because germination and seedling establishment require high energy input and the supplying of carbon backbones for biosyntheses [1,2]. Lipid content in rapeseed is in the order of 42% seed weight and can range in other oilseeds from about 20% seed weight in soybean (Glycine max) to 60–76% in some subtropical species, e.g., sesame (Sesamum indicum) [3]. Even non-oilseed species, such as pea (Pisum sativum L.), contain about 2% of their seed weight as neutral lipids [4].
The vast majority of seeds store lipids in the form of triacylglycerols (TAGs) of various acyl chain lengths and functionality ([5–8]; see [8] in this issue for structural features of TAGs), the desert shrub jojoba (Simmondsia chinensis) being the only known plant storing a long-chain liquid wax in its seeds [6]. Diacylglycerols [9], sterol esters (SE) [10] or tocopherols (TC, vitamin E) [11], phospholipids (PL) and proteins have also been found in seed lipid bodies. Using electron microscopy, Yatsu and Jacks [12] first proposed a structural model for lipid bodies in which the TAG matrix is surrounded by a monolayer of phospholipids with their head group exposed to the cytosol and the acyl moieties facing inward to interact with TAGs, a model that was later confirmed by other studies [13,14]. The most abundant phospholipids are in the order phosphatidylcholine [15,16], phosphatidylserine, phosphatidylethanolamine, phosphatidylinositol and phosphatidic acid. This reflects the composition of the endoplasmic reticulum (ER) from which the lipid bodies are thought to originate, despite some controversy on the exact mechanisms [10,17]. Very recently, Czabany [18] elegantly demonstrated that the lipid core of yeast (Saccharomyces cerevisiae) lipid particles possesses an original organization. TAGs are more or less randomly packed in the center of lipid particles, while SE formed several ordered shells below the surface of the PL monolayer. Such structural arrangement is certainly crucial for lipid mobilization, and will have to be examined for plant lipid bodies as well as its consequences in terms of biogenesis and biodegradation.
Only a limited number of proteins are specifically associated with plant seed lipid bodies. However, this number varies considerably among plant species. There are three in castor bean (Ricinus communis) [19], five in sesame [20,21], eight to nine in Arabidopsis (Arabidopsis thaliana) [2,22], and up to 11 to 20 in rapeseed [16,23]. They all belong to three major groups comprising structural proteins (oleosins and caleosins), enzymes (e.g., steroleosin and lipases) and minor proteins (e.g., aquaporins). In this review, we will consider only the structural proteins and enzymes.
Oleosins, as well as TAG and phospholipid biosynthetic enzymes, are synthesized during seed maturation on the endoplasmic reticulum. The ER origin of seed lipid bodies is nowadays broadly accepted [5], following initial evidence provided by the work of Frey-Wyssling et al. [24] showing that lipid bodies arise by vesiculation. Specialized ER domains enriched in TAG biosynthetic enzymes could channel acyl groups toward TAG formation rather than to the production of membrane lipids [5,25–27]. Newly synthesized TAGs would accumulate until a critical ratio PL/TAG (ca. 3% mol/mol) is reached [28]. Accumulation of TAGs between the two leaflets of the ER membrane leads to the formation of a defined microdomain. The enlarging of the microdomain volume is followed by the budding of growing lipid bodies from ER in the cytoplasmic space. Finally, the nascent lipid body, covered by a monolayer of phospholipids, is released into the cytoplasmic space, and probably undergoes a cycle of consequential fusions [29].
Cytoplasmic lipid bodies can be found in almost any plant cell and tissue. In seeds, storage lipid accumulation strongly depends on seed and silique developmental stage [5]. In seeds undergoing desiccation by the end of seed maturation (orthodox seeds; see for reviews [30] and [31] in this issue) a single cell may contain from dozens to hundreds of lipid bodies of only diameter [5,32]. In marked contrast, non-desiccating seeds (recalcitrant seeds) contain numerous larger lipid bodies [5,15]. The larger diameter of lipid bodies found in recalcitrant seeds is usually explained by a lower amount of oleosins when compared with lipid bodies isolated from orthodox seeds [15]. In albuminous dicotyledonous seeds the site of lipid body accumulation is mainly the endosperm [33]. In exalbuminous dicotyledonous seeds as rapeseed and sunflower lipid bodies are found mainly in the cotyledons and axis of the embryo [34–36]. In seeds from monocotyledonous plants lipid bodies may be located in the embryo scutellum [37] and/or in the aleurone layer [38].
Storage lipids are mobilized rapidly during seed germination and early seedling establishment [39,40]. Several seed lipases have been reported, in castor bean [19] and in Arabidopsis [2]. Both are associated to lipid bodies and shown to hydrolyze TAGs. However, only the lipase from Arabidopsis was clearly demonstrated to be involved in lipid mobilization during seed germination. This raises the question of the existence and role of the other seed lipases.
2 Lipid body integral proteins
The most abundant structural proteins of seed lipid bodies are oleosins, which represent up to 0.4 to 1.6% of the seed weight [35], and even as much as 10% of total seed proteins in sunflower [5].
2.1 Oleosins
Oleosins are a class of mostly alkaline proteins (10–53 kDa) believed to be ubiquitous in oil-storing seeds [35]. The first oleosin was isolated from peanut (Arachis hypogaea) seeds [41] and since then others have been isolated from many higher plant species and also from gymnosperms [5,42]. Seed oleosins may be classified into high and low molecular weight proteins (HMW, LMW) [43,44], which are not immunologically related [45].
Arabidopsis oleosins are encoded by a multigenic family comprising 16 genes, specifically expressed in the floret tapetum (eight genes), in maturing seeds (five genes), or in both maturing seeds and floral microspores (three genes) [46]. Not all proteins have been biochemically identified to date.
The primary structure of oleosins is highly conserved among species. Their organization in three domains consisting of a highly hydrophobic central region flanked by two more polar C- and N-termini [47] largely explains the mechanism of oleosin insertion in lipid bodies. The central hydrophobic domain, which is well conserved across plants, is unique owing to the presence of mostly non-polar amino acid residues [48] comprising in its central part conserved proline residues arranged in the so-called “proline-knot” motif. The embedding of oleosin central hydrophobic region was confirmed by protease protection studies and differential accessibility to antibodies [35,36]. The amphipathic character of the N- and C-terminal domains of oleosin excludes their dipping into the TAG core and instead enables their interaction with the aqueous surrounding. Their length can vary considerably, from nine to 403 amino acids [46]. Conserved charged amino acid residues from the N- and C-termini could interact with the phospholipid head groups at the surface of the lipid body [49]. Most rapeseed oleosins are N-acetylated on their N-terminal alanine residue [50], [Jolivet et al., personal communication]. Moreover oleosins are able to create highly stable oligomeric associations even in the presence of strong ionic detergents as SDS [51–53].
Results concerning the secondary structure of oleosins are contradictory. The oleosin hydrophobic central domain could be formed by two α-helices [54] or two β-sheet structures [55]. The low solubility of oleosins in aqueous media is a roadblock for structural investigations, and to date no satisfactory solvent system has been found. Circular dichroism (CD) spectra of a peptide corresponding to the hydrophobic domain of sunflower oleosin are typical of α-helical structures in SDS micelles, or in 100% TFE, this last solvent notoriously promoting α-helical structures [49]. The fold of this hydrophobic domain, which is almost three times longer than known transmembrane domains, remains unknown while the mechanism of its insertion at the lipid body interface has been more documented [56]. The C-terminal domain comprises predicted amphipathic α-helical structure [47], a motif frequently found within the lipid-binding portion of membrane proteins. Using a modeling program to predict the secondary structure of the N-terminal domain of the 19-kDa rapeseed oleosin, Murphy et al. [57] suggested the possible presence of α-helices in this domain. However, CD and FT-IR spectroscopy measurements performed on a sunflower N-terminal soluble peptide produced in Escherichia coli revealed the presence of mixture of all three possible secondary structures, namely α-helices, β-sheets, and unordered structures [58]. A schematic representation of oleosins within the lipid body interface is shown in Fig. 1.
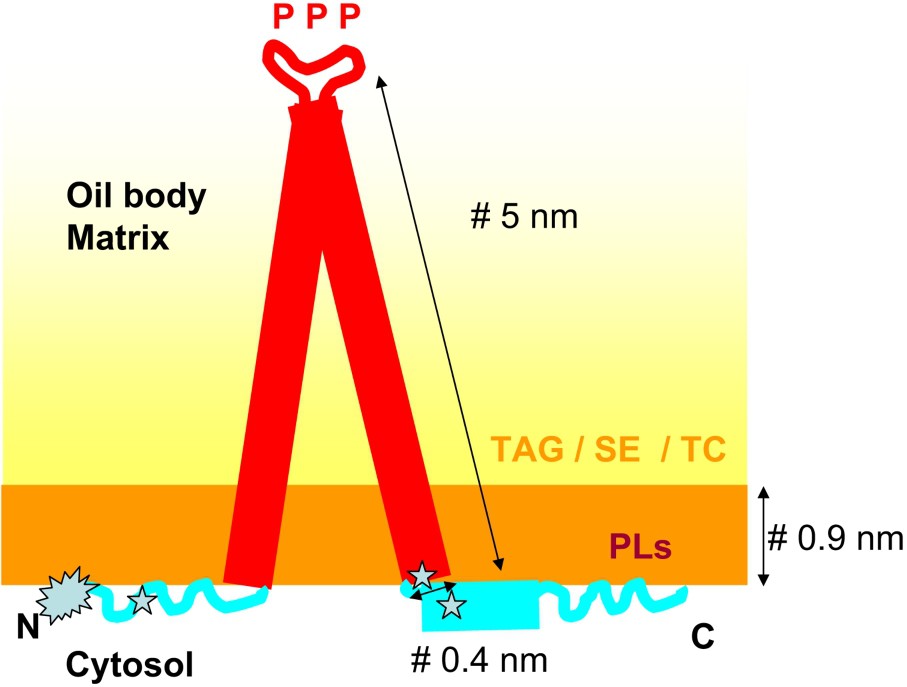
Proposed model of oleosin conformation. The fold of the hydrophobic region embedded in the body matrix is generally described as α-helical, even if an earlier model proposed a β-sheet fold for this region [86]. It contains conserved proline residues (P) involved in protein targeting to lipids. Region enriched with positively charged residues found between the central region and the N and C extensions are presumably involved in interaction with charged PLs () [49]. The N terminus is acetylated () [50].
Investigation of the folding and structure of oleosin in solution remains a challenge, as the proper solubilizing agent remains to be found. FT-IR spectroscopy or alternatively SRCD (Synchrotron Radiation Circular Dichroism) investigation of the structure of proteins in lipid bodies from plants with controlled protein complement or in artificial lipid bodies containing only one type of oleosin will be attractive approaches. The recent demonstration by D'Andréa et al. [59] of the possibility to solubilize oleosins using monophasic aqueous organic solvent mixtures, which previously proved useful for establishing the three-dimensional structure of the membrane domain of subunit β of the Escherichia coli F0F1 ATP synthase [60], is promising.
Oleosins are synthesized on ribosomes and co-translationally inserted into the ER membrane probably via a signal recognition particle [5]. They are exclusively targeted to lipid bodies, even in non-homologous systems as yeast [61–63]. However, the expression of recombinant oleosin genes in either plant or mammalian cell types lacking active synthesis of lipid bodies led to only very low levels of recombinant oleosin protein accumulation on the ER membrane [63]. These facts suggest that oleosins themselves contain the information required for their correct targeting to lipid bodies and that in the absence of suitable lipid bodies they are most presumably unstable and degraded.
The importance of the central hydrophobic domain of oleosins for their correct insertion into lipid bodies was addressed by Van Rooijen and Moloney [64]. These authors investigated the incorporation into rapeseed lipid bodies of a mutated Arabidopsis oleosin lacking one of its three structural domains. The data showed that removal of the last 48 amino acids of the C-terminal domain of the oleosin had no effect on the targeting of the protein in rapeseed seeds. In contrast, deletion of either the N-terminus or the central hydrophobic region strongly impaired targeting of the protein to the lipid bodies, associated with a large reduction in protein accumulation. Site-directed mutagenesis of proline residues within the proline-knot motif strongly affected oleosin insertion into lipid bodies [65], revealing the role of the proline knot motif in protein stability and targeting.
Depending on the organism studied, accumulation of lipids and of oleosins has been reported to be either synchronous or time-separated processes. For example, in rapeseed embryos lipids and oleosins are not accumulated synchronously [39,66]. Also, in oat (Avena sativa) oleosins accumulate during kernel development, while lipid accumulation occurs mainly in the very early stage of development [67]. In contrast, in maize (Zea mays) [37] and soybean [25] TAGs and oleosins appear to accumulate concomitantly during seed maturation on the mother plant.
A high surface/volume ratio favoring the action of lipases [68], the proposed physiological role of oleosins is to maintain lipid bodies as small discrete organelles till germination. They prevent lipid bodies coalescence by steric hindrance and electrostatic repulsion, thanks to the net negative charge of their outer coating [69]. Oleosins are present in high amount in the lipid bodies of seeds undergoing desiccation at the end of the maturation phase (orthodox seeds) and rehydration during germination. Therefore, their stabilizing effect on the lipid bodies seems to be most important during the dehydration/rehydration cycle of these orthodox seeds. In agreement with this, oleosin-deficient lipid bodies of desiccation-sensitive seeds proved unstable to drying and rehydration in vivo [15]. Here, lipid bodies retained their integrity during the drying phase of the cycle and only coalesced upon rehydration of the seeds. Siloto et al. [70] observed that an Arabidopsis mutant lacking the major seed oleosin (18 kDa) exhibits embryos containing abnormally large lipid bodies (therefore showing a lower surface-to-volume ratio) together with a decreased ability to store lipids, which results in a delay of seed germination. Maintenance of high surface-to-volume ratios could facilitate access by lipases and thus accelerate TAG mobilization [60]. Interestingly, very recently Shimada et al. [71] demonstrated a role of oleosins in freezing tolerance of Arabidopsis seeds. Using single and multiple mutants for oleosins, these authors found that germination rates were positively associated with oleosin levels in seeds, and that the size of lipid bodies was inversely related to oleosin content. Importantly, this study confirmed the pivotal role of oleosins in lipid body stability and suggested that oleosins increase the viability of overwintering oilseeds by preventing abnormal fusion of lipid bodies during imbibition in the spring time.
Lipid bodies from mature dry seeds were described to be resistant to endogenous lipase attack [72] as well as to digestion by phospholipases A2 and C [13]. In contrast, lipid bodies isolated from seedlings imbibed for at least two days could be degraded by lipase [72]. Purified lipid bodies are substrates for various lipases, including plant lipases [73]. Integrity of lipid body surface is important to control this process, as upon trypsin digestion, the storage lipids become available to lipase attack [13]. Vance and Huang [47] suggested that oleosins may act as receptors for lipases on the lipid body surface, thereby facilitating initiation of TAG hydrolysis during seed germination and seedling establishment. However, the recent data on seed lipases [2,19] raise the question of how lipases are associated to lipid bodies either by docking on oleosins or by direct insertion in PL monolayer, and how they are activated.
2.2 Caleosins
Caleosins constitute a minor group of integral lipid body proteins found in a wide range of plants, in lipid-accumulating fungi and in single-cell algae [74,75]. They share with oleosins a three-domain structure and their molecular weight is comprised between 25 and 35 kDa. They also possess a half-calcium binding site. Naested et al. suggested a classification of caleosins into three groups based on their structure and expression profiles [75]. This classification was later on updated by Hanano et al. according to the location of the hydrophobic region [76].
Chen et al. demonstrated the capacity of purified recombinant caleosin to stabilize artificial lipid bodies [77]. The role of the hydrophobic central region in anchoring caleosins to lipid bodies was confirmed by “peeling” techniques using digestion of lipid bodies by proteinase K [77,78], and by a study of caleosin mutants lacking various structural domains and sub-domains of the protein [79]. Chen et al. also pointed out the importance of the negative charges at the lipid bodies surface for correct targeting of the protein [77]. A structural model for caleosin insertion inside lipid bodies suggests the importance of amphipathic α-helices in the central hydrophobic region of the protein. The extent of α-helices in an Arabidopsis caleosin (AtCLO1), as deduced from a CD study of the proteins in a set of aliphatic alcohol solutions, generally increases when the medium polarity decreases [80]. Post-translational modifications of caleosins have also been documented. Thus, sesame caleosin was shown to be N-acetylated [50] while Arabidopsis caleosin can be partially phosphorylated at a highly conserved protein kinase CK2 phosphorylation site [80]. Acetylation has been generally hypothesized to stabilize proteins by impeding their turnover [50].
Interestingly, caleosins were found to contain a single highly conserved EF-hand motif located in the N-terminal region [74] and that corresponds to a half-calcium binding site. In agreement with this, Frandsen et al. [81] demonstrated that the EF hand motif of a recombinant rice caleosin is capable of binding calcium. Sesame and Arabidopsis caleosins (AtCLO1, AtCLO3) proved also able to bind calcium [74,82,83] and calcium was shown to strongly affect AtCLO1 solubility and interfacial properties [83].
Poxleitner et al. [84] demonstrated that during germination caleosins are involved in the interaction of lipid bodies with vacuoles and thus play a role in the mobilization of storage lipids. Also, Hanano et al. [76] reported that recombinant Arabidopsis and rice (Oryza sativa) caleosins expressed in yeast exhibit oxidoreductase activity, calcium being crucial for peroxygenase activity. Hence, caleosins could possibly be involved in the biosynthesis of phytooxylipins and thus in plant defense responses. Liu et al. [85] have studied the cellular localization, subcellular distribution and dynamics of two barley (Hordeum vulgare) caleosin isoforms using GFP-fusion proteins. Contrary to oleosins, which are directly associated with lipid bodies, GFP-caleosin first accumulated in the endomembrane system as small vesicles, which moved rapidly and appeared to aggregate and degrade. The existence of an ER-associated form was confirmed for other caleosins [75,81]. Ca2+ is capable to modify caleosin interfacial properties [83], and to mediate aggregation of either purified lipid bodies [86] or artificial lipid bodies [Purktkova et al., unpublished data].
2.3 Steroleosins
Seed lipid bodies from sesame and Arabidopsis also contain enzymes found in minute quantities, which belong to the SDR (short chain dehydrogenase reductase) super family. They have been called sterol dehydrogenases or steroleosins. These enzymes are able to oxidize estradiol and corticosterone, thus exhibiting 17 β and 11 β-hydroxysteroid dehydrogenase activities. Sesame Sop2 can use both NAD and NADP whereas sesame Sop 3 and Arabidopsis HSD1 are active only in the presence of NADP [20,21,87]. HSD1 is also capable to catalyze the reductase reaction using NADPH as co-substrate [87]. Although the physiological substrate of these enzymes remains unknown, a recent report [88] suggests that HSD1 would be involved in regulating growth and development in plants, likely promoting or mediating brassinosteroids effects. Thus, this gene has significant potential for improving growth and yield of canola (which are cultivars of rapeseed variants from which low erucic acid rapeseed oil and low glucosinolate meal are obtained; hence the name “canola” as derived from “Canadian oil, low acid”), and other agricultural crops [88].
3 Biotechnological applications of lipid bodies-associated proteins
Lipid bodies and oleosins are increasingly becoming targets for biotechnology. Lipid bodies can serve as adequate carriers for hydrophobic molecules such as neutraceutical compounds (e.g., antioxidant metabolites), pharmaceutical drugs (e.g., steroid hormones or immunosuppressant drug cyclosporine A), and be incorporated into cosmetic preparations (patent FR2119219). Lipid bodies were also successfully tested as a biocapsule for encapsulating lactic acid bacteria in dairy products [89]. Oleosins have been used as a vehicle for the over-expression in transgenic plants of a variety of fusion proteins including insulin [90], β-glucuronidase [91], xylanase [92], hirudin [93] and cystatin [94]. Oleosin targets the fusion protein to lipid bodies and so allows quick, easy and economically reasonable purification procedure [95]. This technology is currently being developed for full-scale commercial production of a range of recombinant proteins. The leader in this industry is a Canadian company Sembiosys (http://www.sembiosys.com/), which focuses on the lipid body/oleosin technology and its utilization in preparation of drugs for treatment of cardiovascular and metabolic diseases.
4 Concluding remarks and perspectives
The structure and function of lipid-body associated proteins remain largely unknown despite their importance for lipid body biogenesis and abundance. There is a lot to be learned as to the various roles that these proteins play both in structural (stabilization and dynamics) and functional (isoforms, enzyme activities, storage lipid deposition) terms. The establishment of original solvent systems enabling the solubilization of hydrophobic proteins together with promising techniques such as SRCD will help deciphering their structure. Studies on functional roles will require an array of complementary approaches, including cell biology for subcellular localization during biogenesis, molecular genetics for tissue localization, genetics for in planta function, and biochemistry for enzyme activities.
This increased knowledge could be used for biotechnological applications. For example, modulating oleosin levels in oilseeds could have an effect on storage lipid extractability by modifying oilseed behavior during pressing under milder conditions. This possibility could better preserve the quality of residual cake proteins used in animal feed (solubility, digestibility) and increase their value. This is especially true for rapeseed as its economical value (as to July 2008) is based on oil (>1028 €/t) and cake (>212 €/t) prices.
Acknowledgements
Zita Purkrtova was recipient of a joint scholarship from the French Ministry of Foreign Affairs (Egide) and the Ministry of Education, Youth and Sports of the Czech Republic (MSM 6046137305). Our investigations on oil bodies are supported by ANR Génoplante.


