1 Introduction
The development of the seed proceeds through embryogenesis and seed filling, and terminates with entry into a quiescent state that permits seed dispersal and survival for many years in various environmental conditions. The success of germination and early plant growth is largely determined by the physiological and biochemical features of the seed. Of key importance to this success are the reserves stored in the mature seed. These reserves, which are principally proteins (referred to as storage proteins), oils (often triacylglycerols) and carbohydrates (often starch), accumulate during seed filling, withstand desiccation during late maturation, and are used as an initial energy source in the heterotrophic growth phase of germination and seedling establishment [1]. In addition to their central position in the higher plant life cycle, seeds such as those of legumes and cereals are major food sources. In cereal grains, the endosperm serves as the major filial storage tissue, being rich in starch but possessing a protein content of less than 20%. In contrast, the principal storage organs of grain legumes are the embryonic cotyledons (see Fig. 1 of the Foreward of this present issue). Unlike cereals, legume seeds are large, rich in protein, and thereby used as protein sources for animal feed. Furthermore, soybean (Glycine max) is currently the most important source of edible oil.
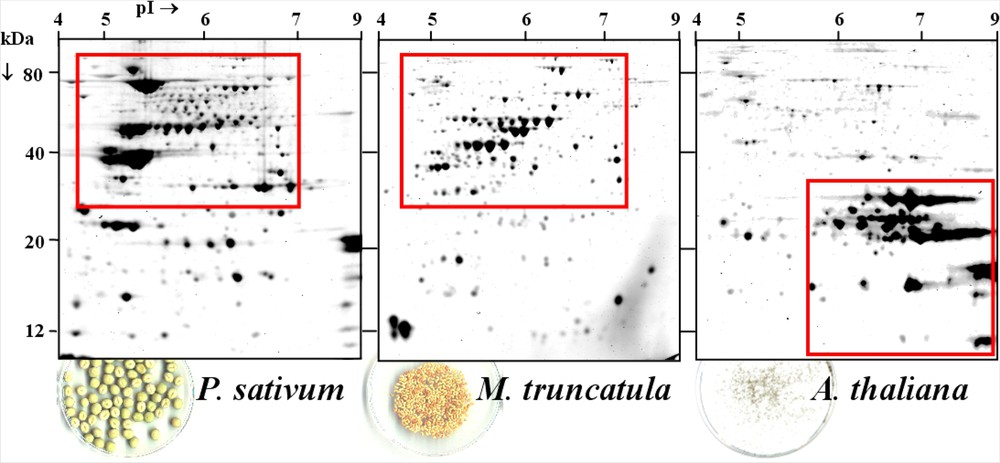
Comparison of mature seed proteomes from pea, M. truncatula and Arabidopsis. The figure shows 2-D gels of total soluble proteins from dry mature seeds of pea, M. truncatula and Arabidopsis. Isoelectrofocusing was carried in an immobilized nonlinear pH 3 to 10 gradient with about 200 μg protein. Proteins were then separated according to size by SDS-PAGE. Proteins were visualized by Coomassie Blue G-250 coloration (for details on protein extraction and separation, see [6,10]).
Because of the ecological, nutritional and economic importance of legume seeds, the biochemical and molecular processes underlying their development have been widely studied (for a review, see [2]). In particular, understanding nutrient uptake, transport, partitioning and metabolism during seed development, and identifying the genetic factors involved have been the focus of much research in recent years in both model plants and crops. Amongst the most common legumes used to study seed reserve accumulation are pea (Pisum sativum), faba bean (Vicia faba), soybean, lotus (Lotus japonicus) and the annual barrel medic (Medicago truncatula). The model legumes M. truncatula and L. japonicus were chosen for large-scale genome sequencing programs because both have relatively small genomes [3]. The synteny assessment between model and legume crops facilitates cross-reference of genomics resources between these species, which accelerates studies on crops with large genome sizes.
In recent years, a combination of genetics, genomics and post-genomics approaches has been used to determine the mechanisms underlying reserve accumulation in legume seeds, with a particular emphasis on storage protein accumulation. In this review, we summarize these findings, highlight the role of metabolic and regulatory networks in coordinating reserve accumulation, and discuss how this knowledge can support our attempts to engineer legume seed composition for added end-user value.
2 Legume seeds: composition and quality
The protein content of legume seeds ranges from 20% to as much as 40%, depending on the species. Seeds of soybean and M. truncatula exhibit high protein contents, whereas pea seed's major component is starch (50% of dry matter). In mature seeds of several legume species, including soybean and M. truncatula, starch content is very low. This is because starch is degraded during seed maturation, probably to provide carbon skeletons for the synthesis of other compounds. Because legume seeds are relatively rich in proteins, several investigations aimed at dissecting their protein composition [4,5]. Advances in proteomics, including the refinement of two-dimensional gel electrophoresis (2-DE) techniques and the development of sensitive mass spectrometry methods for protein identification, allowed the detailed analysis of protein composition in legume seeds. 2-DE maps are now available for seeds of M. truncatula, soybean and white lupin (Lupinus albus L.) [6–10]. The M. truncatula seed proteome map comprises about 224 identified proteins [10] and a 2-DE proteomics reference map comprising 422 identified proteins has been established from developing soybean seeds [7].
A comparison of protein composition in mature seeds of M. truncatula, P. sativum and A. thaliana is presented in Fig. 1. In the reference 2-DE maps from legume species, including soybean, the most abundant protein spots were detected between 30 and 80 kDa under reducing conditions (Bourgeois M., personal communication, [6–9]). They correspond to the principal storage proteins, namely the 7S (vicilin, convicilin) and 11S (legumin) globulins. In contrast, the Arabidopsis seed proteome is mainly composed of 12S (cruciferin) globulins of molecular weight ranging between 16 and 35 kDa. The striking differences in storage protein composition between legume species and Arabidopsis reinforce the usefulness of developing a legume-specific model to investigate processes controlling seed protein accumulation in these plants. Moreover, the synthesis of seed storage proteins is not developmentally separated in Arabidopsis (AtGenExpress Consortium, [11]), whereas in grain legumes and the model species M. truncatula, the onset of vicilin synthesis precedes that of legumins [6,12], making M. truncatula appropriate for studying legume storage protein accumulation and regulation.
Globulins represent about 80% of total seed protein content. A recent study showed that the average amino acid composition of M. truncatula seed proteins is close to that of pea [13], with relatively high lysine content, an essential amino acid that is limiting for nutritional purposes of human and animals in seeds of many crops, including cereals. However, as for the major storage proteins of barley and maize grains (prolamin, glutelin), the predominant proteins of legume seeds are low in the sulfur-containing amino acids cysteine (Cys) and methionine (Met) (<2%) and in tryptophan (<1%). The two latter also belong to the category of essential amino acids, and their low level restricts the nutritional value of legume seeds as animal feed. The storage protein fractions present in seeds are directly responsible for the unbalanced overall amino acid composition since free amino acids only represent negligible amounts (less than 1%) of total nitrogen in mature seeds [13]. The nature of seed storage proteins is genetically programmed but their rate of accumulation depends on nutrient availability, partitioning and metabolism during seed filling.
3 Metabolic control of seed filling in legumes
Two distinct phases characterize the development of legume seeds: a phase of histo-differentiation associated with a high mitotic activity and a phase of reserve synthesis and accumulation [14]. Nutrients coming from the phloem are downloaded into the seed coat. During the pre-storage phase, a transient accumulation of proteins and starch occurs in this tissue [15] and high activities of seed-coat invertases create a high hexose environment promoting embryo cell division [2]: seed sink strength, which is linked to the number of cotyledon cells, is then determined. At the onset of seed filling, a high rate of sucrose transport into the embryo induces differentiation processes and promotes storage activities [16]. During the storage phase, seed coat-located nutrients are transported to the embryo where they are metabolized into storage compounds. Sucrose synthase activity in the seed coat is associated with starch synthesis in the embryo [17]. Sucrose also induces the transcriptional up-regulation of several enzymes, notably phosphoenolpyruvate carboxylase [2] whose activity correlates with seed protein content in soybean cultivars [18] and in transgenic faba bean plants overexpressing this enzyme in a seed-specific manner [19].
The accumulation of seed reserves results from concerted processes occurring in parallel in the main seed compartments (embryo, endosperm, seed coat). Metabolite transport, transient storage, remobilization occur in the seed coat and the endosperm to fuel in a timely manner the accumulation of storage compounds in the cotyledons. Seed coat sucrose and amino-acid transporters have been isolated in legumes and shown to control the flux of nutrients to the embryo [20,21]. Various proteases were found to be preferentially accumulated in these young maternal tissues [10], probably in relation to the endogenous remobilization of amino acids to the growing embryo. In support of this hypothesis, it has been shown that whole seeds cultured in vitro in the absence of exogenous nitrogen are able to initiate the accumulation of embryo storage proteins by recycling nitrogenous compounds from the embryo-surrounding tissues, whereas isolated embryos are not [22].
With the availability of legume genomic resources, comprehensive catalogs of transcripts and proteins present in the various seed tissues were constructed, which provide a global view of the processes occurring in parallel in these seed compartments. A combined transcriptome and proteome analysis of developing M. truncatula seeds has identified tissue-specific features at the onset of seed filling [10]. For example, a compartmentalization of sulfur assimilation enzymes between seed tissues was revealed that may regulate the availability of sulfur-containing amino acids in embryo cells. These findings suggest that sulfate in the tissues surrounding the embryo is mainly incorporated into glutathione and defence-related metabolites, whereas most of the sulfate entering the embryo is utilized for the synthesis of sulfur-containing amino acids. These results, along with those of Catusse et al. [23] documenting the compartmentalization of metabolic activity between the radicle, cotyledons and perisperm in germinating sugarbeet seeds, are indicative of a metabolic control of seed development and germination through the partitioning of metabolic pathways between seed tissues. In soybean, laser capture microdissection of every seed tissue throughout seed development was performed, and the captured mRNAs were analyzed with an Affymetrix GeneChip [24]. The results indicate that at least 22,000 diverse mRNAs are required for the formation of a globular-stage soybean embryo. Genes specifically expressed in the diverse seed compartments (e.g. endosperm, hilum, suspensor, and embryo proper) were identified, highlighting the specialization of each tissue, which may be controlled by the tissue-specific genetic programs of either maternal (seed coat) or zygotic (embryo) origin, but also by the energy and oxygen status of the different tissues. Due to their location, the embryos initially develop in an environment of low light and oxygen availability [25], which may affect ATP production and biosynthetic activities. As a way of controlling biosynthetic fluxes during maturation, embryos become photosynthetically active [26], providing oxygen and ATP for respiration and biosynthetic activities.
4 Regulatory network underlying seed filling
While the main metabolic pathways necessary for reserve accumulation are well characterized, the exploration of the regulatory networks operating in legume seeds is still in its infancy. The few transcription factors known to regulate seed storage protein synthesis in legumes were characterized in bean (Phaseolus vulgaris). The expression of phaseolin, the most abundant protein of bean seeds, is activated by B3-domain transcription factors (ABI3-like factors and FUSCA3, [27,28]), and repressed by bZIP factors before seed filling (regulator of maturation ROM1, [29]) or during late maturation (ROM2, [30]). Li et al. [27] showed that this transcriptional activation does not occur in the absence of abscisic acid (a plant hormone involved in maintenance of the developing embryo in the maturation program and in the accumulation of seed storage compounds), highlighting the importance of this hormone in the transcriptional regulation of seed reserve synthesis. Furthermore, variations in chromatin structure play a crucial role in regulating storage protein synthesis. For example, activation of the phaseolin promoter requires chromatin remodeling [27]. In this context, it is worth noting that the nuclear proteome of developing M. truncatula seeds contains chromatin-modifying enzymes and RNA interference proteins that play roles in RNA-directed DNA methylation and may be involved in modifying genome architecture and accessibility during seed filling [31].
Recently, transcriptomics experiments have been performed that identified hundreds of regulatory factors differentially expressed during seed filling in M. truncatula [10,32,33], thereby providing a basis for understanding the regulatory networks governing this process. For example, expression profiles of over 700 M. truncatula genes encoding putative transcription factors were analyzed throughout seed development using real-time quantitative RT-PCR [33]. Some of the transcription factors co-expressed with storage protein mRNAs correspond to those already known to regulate storage protein synthesis in Arabidopsis, whereas the timing of expression of others was related to the delayed expression of the legumin-class storage proteins observed in legumes. Some of these transcription factors display an embryo specificity with no obvious orthologs in Arabidopsis (e.g. a protein with a B3 domain, bHLH and bZIP factors), suggesting a specific function in legume seeds. Moreover, a gene expression atlas was recently generated that provides a global view of gene expression in all major organ systems of M. truncatula, including developing seeds [34], offering the opportunity to search for seed-specific regulatory factors. Research focused on these seed-specific transcriptional regulators and their regulation of target genes is essential for gaining further insight into the mechanisms governing reserve accumulation in legume seeds.
5 Intra- and inter-population genetic diversity for seed composition
Genetic variation for seed composition was investigated in legume crops. In a collection of 59 pea lines, large variations in starch (28 to 56%) and protein (14% to 31%) content were observed [4]. On average, the wrinkled seeds showed significantly lower starch content than the lines with other seed phenotypes. This wrinkled seed phenotype is commonly observed in rugosus pea mutants affected for one of the enzymes controlling starch accumulation ([35,36] and references therein). As compared to pea, lower variations in seed protein content, ranging from 37 to 41%, were observed in a collection of soybean cultivars [37]. In soybean seeds, a decline in protein content was observed after decades of selection and breeding, which could be attributed to the fact that, during the past several decades, breeding efforts have emphasized increased oil content.
In seeds of major legume crops, high-protein content was correlated with an increase in the proportion of globulins [38], which are the most abundant proteins present in seeds of legumes. Significant biodiversity in seed globulin composition was observed between legume crops. For example, in soybean the content of 11S (glycinin) globulins is always higher than that of 7S (beta-conglycinin) globulins, whereas 7S vicilin is the most abundant protein of pea seeds. Moreover, in comparison with soybean, pea lines show more variation in seed globulin content, enabling a wider range of applications. For instance, the 7S/11S globulin ratio ranges from 1.2 to 8 in pea, whereas it only ranges from 0.5 to 0.8 in soybean [4,39]. In pea, this ratio is a good indicator of seed protein content, since it was found to increase in seeds with lower protein content [40].
Sequence comparisons and crystal structures of 7S and 11S globulins make it clear that the two classes of proteins share common ancestry [37,41]. The degree of divergence in the DNA sequence of the genes encoding 11S glycinin and 7S beta-conglycinin in ancestral and modern soybean cultivars has been investigated [37]. Overall, the Southern hybridization patterns were similar among the ancestral cultivars and those derived from them, suggesting a high degree of conservation of seed-storage protein genes. In addition, a high degree of similarity in quantitative and qualitative accumulation of seed storage proteins was observed among varieties (ancestral and modern cultivars) of soybean [37] or alfalfa (Medicago sativa L., [42]), suggesting a high degree of uniformity in seed filling. However, in a collection of 50 lines of M. truncatula, electrophoretic analyses of total seed protein extracts revealed 46 major polypeptides, of which 26 were polymorphic within the collection [43]. Lines contrasting for globulin profiles were identified to allow for the genetic determination of seed storage protein accumulation, which will be relevant for legume crop improvement.
6 Environmental and genetic determinants of seed reserve accumulation
Seed reserve accumulation is not only determined by the embryo's intrinsic capacity to accumulate storage compounds, but also by processes occurring in other plant parts during the life cycle, which are controlled by both genotype and environment. Of key importance for reserve accumulation is the supply of nitrogen and carbon acquired by roots, and their remobilization from almost all vegetative organs during seed filling [44,45]. In leaves, nitrogen remobilization is associated with the degradation of the photosynthetic apparatus [46], which, in turn influences carbon fixation. Nitrogen and carbon are imported into developing seeds through the phloem, mainly in the form of organic compounds (e.g. sugars, glutamine and asparagine) [2]. Although seed weight and protein content are mostly determined by the availability of assimilates (source strength), variability in sink strength (assimilate demand) also plays an important role [47]. Sink strength is determined by the ability of the sink to transport and supply the embryo with amino acids and sucrose [2]. Sulfur nutrition also influences seed composition, for instance by modifying the legumin/vicilin ratio [48]. Sulfur-containing compounds supplied to the seed have been identified (sulfate, S-methylMet and glutathione) [49,50] and their importance for seed reserve synthesis is being studied along with their transport and delivery to storage organs [51].
In a recent study, quantitative trait loci (QTL) for seed traits and indicators of sink strength and source nitrogen capacity were searched to identify the genetic factors responsible for seed protein content and yield fluctuation in pea [47]. Remarkably, most QTL for seed traits and plant source capacity mapped to clusters in the genome. The underlying genes may have pleiotropic effects on source-sink relationships. In most environments, the Le and Afila genes, which control internode length and the switch between leaflets and tendrils respectively, determine plant nitrogen status. Depending on the environment, these genes were linked to QTL for seed protein content and yield, suggesting that source-sink relationships also depend on growing conditions. Clusters of QTL for seed traits only were also detected that may correspond to genes specifically involved in seed development and metabolism. These genes may control processes determining sink strength and/or the rate of assimilate accumulation in pea seeds. A recent study analyzed the role of the PA2 locus in the control of the quantity of this storage albumin and in the control of polyamine metabolism [52].
7 Toward genetic improvement of legume seed quality
Improving the nutritional quality of legume seeds is a desirable goal. For example, increasing and stabilizing seed protein content while maintaining seed yield is an important challenge for pea breeding. Such improvements might be achieved by increasing nitrogen uptake by roots, as well as nitrogen remobilization and/or translocation from vegetative parts. Modification of the embryonic control of nutrient uptake or metabolism may also be a strategy for increasing the seed protein content. For example, seed-specific expression of a bacterial phosphoenolpyruvate carboxylase in Vicia narbonensis increases protein content and improves carbon economy by inducing a shift of metabolic fluxes from sugars/starch into organic acids and free amino acids [19]. However, the number of seeds per pod decreased in these transgenic plants, which was compensated by an increase in seed dry weight with no consequence on seed yield. As a second example, seed-specific overexpression of the amino acid permease VfAAP1 in pea was shown to increase amino acid supply, seed nitrogen and storage protein content [53], indicating a stimulation of seed protein synthesis by increased amino acid availability.
In addition, as legume seeds are deficient in sulfur-containing amino acids (among which Met is essential), an ambitious aim in plant breeding is to specifically increase their content in seeds of major legume crops. Toward this goal, several strategies have been employed (see [54] for a review). As an example, the sulfur-amino acid content in seeds of chickpeas (Cicer arietinum L.) could be increased by expression of a recombinant sulfur-rich sink protein [55]. In a recent study, several QTL controlling Met and Cys contents in soybean seeds were detected [56]. These QTL along with those controlling seed protein content [47] provide important information to breeders targeting improvements in the nutritional quality of legume seeds. Natural variation can also be exploited to isolate novel genes or alleles in pea germplasm collections (for example, http://193.50.15.18/legumbase/; http://www.jic.ac.uk/germplas/pisum/; www.ars-grin.gov/). Finally, the method for Targeting Induced Local Lesions In Genomes, or TILLING, has recently been set up in pea (http://urgv.evry.inra.fr/UTILLdb; [57]) and soybean (http://www.soybeantilling.org/; [58]), which in addition to providing screening resources for isolating different mutant alleles corresponding to genes of interest with known sequences, allows the improvement of crop plants since favorable alleles detected in TILLING mutant collections can be used directly in selection.
Acknowledgements
We thank J. Verdier, M. Bourgeois, and H. Zuber for providing useful information regarding seed composition and regulation.


