1 Introduction
Wheat is a primary food crop that is planted widely around the world (USDA 2003). However in many wheat-planted areas of the world, abiotic stresses such as water shortage, cold, heat, salt, etc. often limit wheat productivity [1]. Consequently it is of great importance to understand the molecular adaptation mechanisms of organisms to abiotic stresses. This understanding could contribute to our ability to strengthen and improve abiotic stress tolerance in wheat.
NADP-ME is a ubiquitous enzyme and catalyzes the oxidative decarboxylation of (L)-malate and NADP to produce pyruvate, CO2 and NADPH in the presence of a bivalent cation [2–9]. Plant NADP-MEs are divided into photosynthetic NADP-MEs and non-photosynthetic NADP-MEs according to their physiological roles [5,6,10]. The former exist in C4 bundle sheath chloroplasts and cytosol of CAM plants, where they supply CO2 to ribulose-1,5-bisphosphate carboxylase/oxygenase for carbon fixation. Their expression is regulated by light. Although abundant in leaves, photosynthetic NADP-MEs are expressed in vegetative tissues [11–13]. The photosynthetic NADP-ME gene from maize in transgenic tobacco is reported to reduce the stomatal conductance and further enhance water use efficiency (WUE) in transgenic tobacco [14]. The basic physiological definition of WUE equates to the ratio of photosynthesis to transpiration, also referred to as transpiration efficiency, which describes the relationship between water use and crop production and is measured as the biomass produced per unit transpiration [15–31]. This may provide additional insight into the photosynthetic role of NADP-ME gene in the control of plant WUE [32–43].
The non-photsynthetic NADP-MEs, in contrast, are found in plastids or cytosol in various plants and have played many important roles in their physiology. Functions include regulating guard cell osmotic pressure and stomatal movement, maintaining cellular pH and ionic balances, ripening the fruit, the fatty acid biosynthesis by supplying both carbon and NADPH and the synthesis of organic acids [16–19,44–53]. Plant NADP-MEs can therefore be divided into plastidic isoforms and cytosolic isoforms according to their subcellular location. Recently the non-photosynthetic NADP-MEs have been associated with plant defensive responses [3,20,21]. For example, NADP-ME is conducive to synthesis of lignin, flavonoid and activated oxygen species after being induced following wounding or pathogen attacks or UV-B radiation [20,22,23].
Because it is such an important enzyme, NADP-MEs have been well studied in C4 and CAM plants [5–7,11,24–28,42–47]. In addition, cytosolic and plastidic NADP-ME isoforms have also been reported from C3 plants such as rice, tobacco and, Arabidopsis thaliana and others. In wheat, in situ immunolocalization indicates that there is one 72 kDa isoform in chloroplasts of the mesophyll cells [29]. What is more, NADP-ME activity increased after the treatment of wheat leaves with cellulase and macerozyme [30]. But little is known about the wheat NADP-ME genes. Here, we first cloned and analyzed the putative wheat NADP-ME genes, and determined the response of wheat NADP-ME genes to a wide variety of stimuli [48–53].
2 Materials and methods
2.1 Plant growth and stress treatments
Wheat (Triticum aestivum L) cultivar (Shijiazhuang 4185) was grown in vermiculite in a culture room at 25 °C with a 12 h light and 12 h dark cycle. Abiotic stress treatments were applied to 2-week-old seedlings during the photoperiod for specific time periods and control was performed without treatment and named 0 h. Salinity, Polyethylene glycol (PEG) 6000, ABA and SA treatments were applied by submerging the roots of the plants in a Hoagland solution containing either 200 mM NaCl or 20% (w/v) PEG or 200 um ABA or 3 mmol/l SA during the photoperiod. Low temperature treatments were applied by transferring plants to 4 °C environment. Dark treatments were applied by covering plants with a black cloth during a light period. Treated groups were harvested at 3, 6, 12 and 24 h after treatments, with the exceptions of the dark treatment groups, which were harvested at 6, 12, 24 and 48 h after treatment. All tissues harvested for RNA extraction were weighed, immediately frozen in liquid nitrogen, and stored at .
2.2 RNA extraction and the first-strand cDNA synthesis
Total RNA was extracted from the experimental wheat plants using RNArose Reagent method (Watson Biotechnologies, Inc., Shanghai, China). The quantity and quality of total RNA was measured by spectrophotometer (NanoDrop ND-1000, America) and 1% agarose/EtBr gel electrophoresis. One microgram of the isolated total RNA was used to synthesize first-strand cDNA using AMV reverse transcriptase according to the instructions of the manufacturer (Takara, Japan).
2.3 Cloning of partial TaNADP-ME1 and TaNADP-ME2 genes
The reported nucleotide sequence [NCBI: AY3158220] of maize non-photosynthetic NADP-ME and the nucleotide sequence [NCBI: AY435404] of rice, Oryza sativa NADP-ME were respectively selected as query sequences. We searched with these in a wheat expressed sequence tag (EST) database (http://www.ncbi.nlm.nih.gov/Blastn) by using the blastn (nucleotideBlast) program. Eighty-seven and one hundred homologous wheat ESTs were retrieved. Five overlapping ESTs [NCBI: CJ689555, NCBI: CV763610, NCBI: CJ706946, NCBI: CJ617525, NCBI: BJ290869] were assembled into a contig without a complete open reading frame (ORF) and named TaNADP-ME1. Five other overlapping ESTs [NCBI: CJ952974, NCBI: CJ667871, NCBI: CJ801001, NCBI: CJ906404, NCBI: CJ960929] were assembled into a contig with a complete open reading frame (ORF) and named TaNADP-ME2. Specific primers (Table 1) for TaNADP-ME1 (PME1-5, PME1-3) and for TaNADP-ME2 (PME2-5, PME2-3) were designed and used to amplify the cDNA of TaNADP-ME1 and TaNADP-ME2 from the untreated leaf tissues. PCR reaction was performed as follows: 94 °C for 2 min; 30 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 2 min; and finally 72 °C for 10 min. The corresponding band was recovered, subcloned into pMD18-T vector (Takara, Japan) and sequenced.
The primers used in this study.
| Primer | Sequence (5′-3′) |
| PME1-5 | 5-CGCAACCAACTCAACTCACC-3 |
| PME1-3 | 5-GCAAGAACAGCGACAGACAAC-3 |
| PME2-5 | 5-GCCGAGAGTTTGAGGAGATG-3 |
| PME2-3 | 5-GCATATGGGGGAAGGAGATT-3 |
| E1GSP1 | 5-ACTCCTGGAGGAAGAAGGCCACGCA-3 |
| E1NGSP1 | 5-CGCATCCCACTCCCTCTCGGTAAAG-3 |
| E1GSP2 | 5-CGGAAACACTAGCTGCACAGGCCACAA-3 |
| E1NGSP2 | 5-CAGAAAGATATCTGCCCTCATCGCTGCTAG-3 |
| E2GSP1 | 5-AGGGGCACGGTGTAACTGCGGAG-3 |
| E2NGSP1 | 5-TCCTGGTGCTCCTGGGAGACGATG-3 |
| E2GSP2 | 5-GCCTACGTGTTCCCCGGCTTCG-3 |
| Be1-5 | 5-GAAGCATACAAATGGACCAAGG-3 |
| Be1-3 | 5-CAAGAACAGCGACAGACAACAA-3 |
| Be2-5 | 5-GTGGAGTACGAGGGGAAAAC-3 |
| Be2-3 | 5-GCATATGGGGGAAGGAGATT-3 |
2.4 5′-RACE and 3′-RACE for TaNADP-ME1 and TaNADP-ME2 genes
5′-RACE and 3′-RACE for TaNADP-ME1 and TaNADP-ME2 genes were performed because the isolated cDNAs (TaNADP-ME1 and TaNADP-ME2) were half-baked (partial). 5′-RACE and 3′-RACE were carried out according to the manual of the manufacturer (Clontech). Gene specific primers were designed according to the partial cDNAs sequence. They are as follows: E1GSP1,E1NGSP1, E1GSP2, E1NGSP2, E2GSP1, E2NGSP1 and E2GSP2, as shown in Table 1. PCR reaction was performed for 94 °C 2 min; ten cycles of 94 °C for 30 s, 65 °C for 30 s, 72 °C for 2 min; 30 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min; and finally at 72 °C for 10 min. The diluted primary PCR product was used as template in a nested PCR reaction. The nested PCR conditions were same as in the initial reaction. The products were run on a 1% agarose gel containing EtBr and the corresponding DNA bands were purified and subcloned into the pMD18-T vector (Takara, Japan) and sequenced.
2.5 The gene expression patterns of the two NADP-ME genes in different tissues and under various abiotic stresses
In order to identify the expressions of the two genes in different wheat tissues without treatment and in leaves under different abiotic stresses, the semi-quantitative RT-PCR was performed. Total RNA was extracted from different wheat tissues without treatment and from leaves with different treatments (as mentioned above). RNA samples were digested with DNase I (RNase Free) at 37 °C for 1 h. Then DNA-free RNA was reverse transcribed with Oligo(dt)18 primer and PrimeScript Reverse Transcriptase at 42 °C for 1 h (Takara, Japan). Because the two genes are less similar in the 3′ untranslation region, gene specific primers for semi-quantitative RT-PCR were respectively designed in this region. They are as follows: Be1-5 and Be1-3 (Table 1) for TaNADP-ME1, Be2-5 and Be2-3 (Table 1) for TaNADP-ME2. The linear range of amplification was determined to establish the number of cycles for semi-quantitative RT-PCR. Finally the cDNAs were amplified with gene specific primers. The primers for wheat actin were used with an internal standard. In order to rule out the possible contamination from genomic DNA, a control reaction was completed using non-reverse transcribed RNA as a template.
2.6 Sequence analysis and phylogenetic tree
Nucleic acid and amino acid sequence analysis were done using DNAMAN sequence analysis programs (DNAMAN version 5.2.2). Prediction of putative subcellular localization was carried out using the WoLF PSORT on-line program (http://wolfpsort.org) and the TargetP program (http://www.cbs.dtu.dk/services/TargetP). The prediction of signal peptide and putative cleavage sites were completed using the ChloroP1.1 Server on-line program (http://www.cbs.dtu.dk/services/ChloroP). The putative secondary structure is predicted by the scratch protein predictor program (http://scratch.proteomics.ics.uci.edu). Multiple alignment of the complete amino acid sequences of NADP-ME of different plants accessible from public databases was performed using the CLUSTALW multiple alignment program (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The phylogenetic tree was constructed with the neighbor-joining (NJ) method using the mega 4.0 program [42]. Bootstrap analysis was computed using 1000 replicates and excluding positions with gaps.
3 Results
3.1 Isolation of TaNADP-ME1 and TaNADP-ME2 cDNA clones
To obtain the full-length cDNA sequence, bioinformatic analysis and RACE-PCR techniques were employed. As a result, full-length cDNAs of TaNADP-ME1 and TaNADP-ME2 were obtained. Complete TaNADP-ME1 gene [NCBI: EU170134] includes 2392 bp and has an open reading frame (ORF) of 1944 bp with 148 bp and 300 bp segments corresponding to the 5′ and 3′ untranslated regions. The TaNADP-ME1 gene encodes 647 amino acids with a theoretical molecular mass of 71 kD and pI of 6.5. The TaNADP-ME2 gene [NCBI: EU082065] contains 2210 bp and has an open reading frame (ORF) of 1713 bp with 164 and 333 bp segments corresponding to the 5′ and 3′-untranslated region. TaNADP-ME2 gene encodes 570 amino acids with a theoretical molecular mass of 63 kD and pI of 5.6. The TaNADP-ME1 and TaNADP-ME2 are rich in Ala, Glu, Gly, Leu and Val. Further research with the scratch protein predictor showed that these two putative proteins in secondary structure are abundant in α-helixs. The similarity of these two genes in whole amino acids is 73.26% and in nucleotide sequences are 64.08%. Their 3′-end untranslated region include a polyadenylation signal and a poly (A) tail. According to WoLF-PSORT (http://wolfpsort.org/) and TargetP program (http://www.cbs.dtu.dk/services/TargetP/), TaNADP-ME1 and TaNADP-ME2 genes are located in chloroplasts and the cytosol, respectively. Further research with ChloroP1.1Server (http://www.cbs.dtu.dk/services/ChloroP/) indicates that the predicted signal peptide of TaNADP-ME1 has a length of 45 amino acids and a cleavage site location between R45 and A46.
Previous research has reported that five conserved sites exist in known plant NADP-MEs [6,21,31,32]. The first conserved site (Site I) is VYTPTVGEACQKYG, the second site (Site II) is IQVIVVTDGERILGLGDLGCQGMGIPVGKL, the third (Site III) and fourth conserved site (Site IV) are QFEDFANHNAF and FNDDIQGTASVVL, the last site (Site V) is LFLGAGEAGTGIAEL. Among them, Sites I, II and V are NADP-binding sites. The functions of Sites III and IV remain unknown. The two NADP-ME genes contain these five highly conserved sites, while the conserved I228 is substituted by T in Site II of TaNADP-ME1, and T121 is altered to V in Site I of TaNADP-ME2. These alterations in wheat are unique in reported plant NADP-MEs. Whether these changes might lead to variations in enzyme activity needs further study.
According to the multiple sequence alignment with the deduced amino acid sequence of known plant NADP-ME genes (Fig. 1), the two NADP-MEs sequences discussed here are highly homologous to other plant NADP-MEs, as seen by the rather high identity percentages with other plant NADP-MEs. The isoform TaNADP-ME1 shares identity of 89% with OschlME6, 85% with Zmno-pME, 84% with ZmchlME and OscytME2, 76% with NtcytME2, 69% with NtchlME, and 67% with ZmcytME. TaNADP-ME2 shares identities of 90% with OscytME2, 84% with OschlME6, 83% with Zmno-pME, 82% with ZmchlME, 75% with ZmcytME, and 78% with NtcytME2 and NtchlME, respectively. In a way, the result of multialignment displays the divergence of plant NADP-ME, which mainly occurs in the N-terminal end of protein.

Sequence alignment of NADP-MEs from different plants. The sequences used in this multialignment, in addition to these two wheat NADP-MEs, are as follows: OschlME6 (Oryza sativa, D16499), OscytME2 (Oryza sativa, AY435404), Zmno-pME (Zea mays non-photosynthetic NADP-malic enzyme, AY315822), ZmchlMe (Zea mays, U39958), NtcytME2 (Nicotiana tabacum, DQ923118), NtchlME (Nicotiana tabacum, DQ923119), LechlME1 (Lycopersicon esculentum, AF001269), LecytME2 (Lycopersicon esculentum, AF001270), AtNADP-ME1 (Arabidopsis thaliana, AY062734), AtNADP-ME4 (Arabidopsis thaliana, AT1G79750), ZmcytME (Zea mays, AJ224847).
3.2 Tissue-specific analysis of TaNADP-ME1 and TaNADP-ME2 genes in wheat
The expressions of these two NADP-ME genes were analyzed in different tissues of 2-week-old seedlings without treatment during the light period from the wheat cultivar, shijiazhuang4185. As shown in Fig. 2A, TaNADP-ME1 is expressed in root, stem, leaf and the expression level of TaNADP-ME1 is obviously higher in leaves than in roots. However, the expression pattern of TaNADP-ME2 is different from TaNADP-ME1. TaNADP-ME2 is expressed in the roots, stems and leaves of wheat and the expression level is almost equal in quantity (Fig. 2B). This study shows that the plastidic and cytosolic isoforms can co-express in the same tissue.
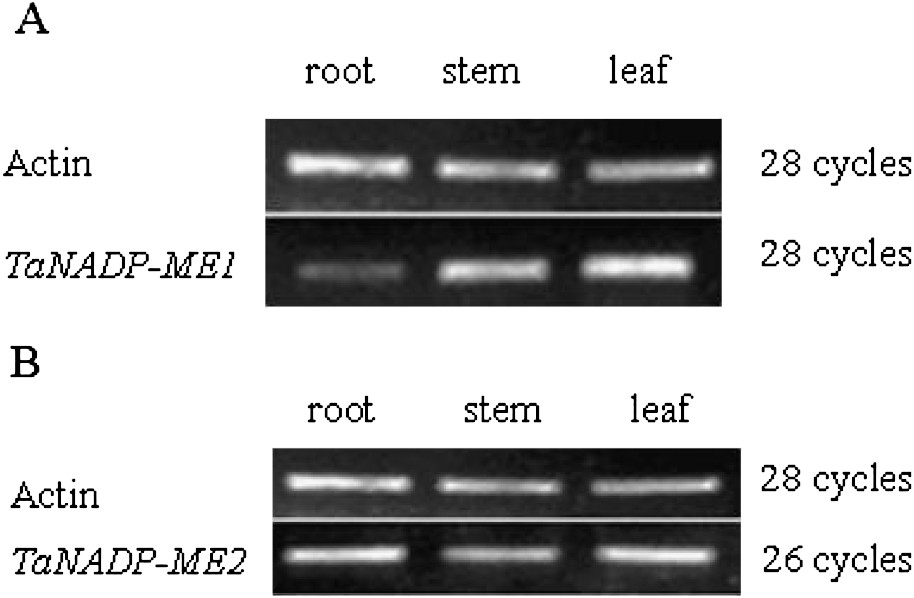
Expression analysis of TaNADP-ME transcripts in different tissues of wheat. DNA-Free total RNA from different tissues (roots, stems, leaves) of 2-week-old wheat seedlings were analyzed by semi-quantitative RT-PCR. The actin (GenBank accession no. AB181991) transcript was used an internal control. Control reaction was performed using non-reverse transcribed RNA as a template in order to eliminate the contamination from genomic DNA. No amplification product was detected in control reactions. A 486-bp fragment of TaNADP-ME1, and a 415-bp fragment of TaNADP-ME2 were amplified by 28 cycles and 26 cycles, respectively. Experiments were repeated at least four times with similar results. (A) Tissue-specific expression analysis of TaNADP-ME1 transcripts. (B) Tissue-specific expression analysis of TaNADP-ME2 transcripts.
3.3 Expressions of TaNADP-ME1 and TaNADP-ME2 under different abiotic stresses or exogenous hormone regimes
Plant responses to abiotic stress are composed of multiple signaling pathways [33]. What is more, various signaling pathways have cross-talk communication [43–47]. In order to further confirm if the TaNADP-ME1 and TaNADP-ME2 are regulated by abiotic stresses and/or exogenous hormones, we investigated their corresponding transcript accumulation in 2-week-old seedlings under various treatments. Control was performed without treatment and named 0 h. The expression levels of TaNADP-ME1 in leaves clearly decreased to the lowest point at 6 h following application of ABA (200 uM), where it was almost not being detected, then began to recover until the 24 h treatment (Fig. 3A). When applied to SA treatment, the expression levels for TaNADP-ME1 was reduced at first 6 h treatment then began to recover until the 24 h treatment (Fig. 3A). With respect to TaNADP-ME2, the expression levels were reduced by ABA and SA treatments (Fig. 3B). For the ABA treatment, the expression amounts of TaNADP-ME2 reach to lowest point at 6 h. In the SA treatment, the expression amounts of TaNADP-ME2 decreased to least at 3 h treatment, then began to ascend till 6 h and again started to descend till 24 h treatment. When treated with 4 °C, NaCl, PEG, TaNADP-ME1 was down-regulated and low temperature treatment was more distinct. For the PEG treatment, the TaNADP-ME1 transcript declined to its lowest level in the first 12 h, then ascended slightly for the duration of the treatment, but levels remained lower than the control (Fig. 3A). Compared with TaNADP-ME1, the expression of TaNADP-ME2 was also down-regulated (Fig. 3B). When exposed to 4 °C, the expression level of TaNADP-ME2 dropped in the first 6 h and then ascended in the rest 18 h, which was lower than the control response. When treated with NaCl and PEG, the transcript accumulation of TaNADP-ME2 continuously decreased during the treatment (Fig. 3B).
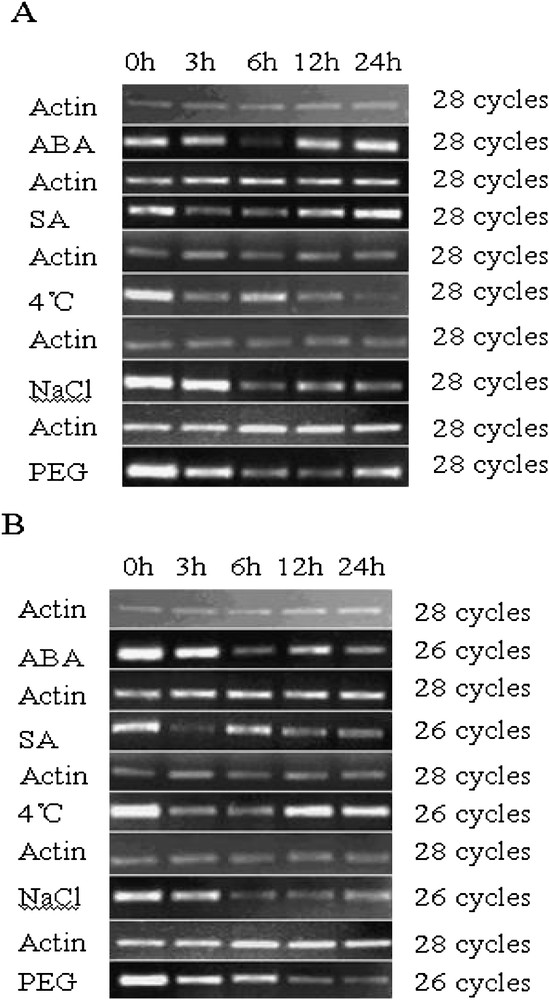
Expression of TaNADP-ME2 in leaves under various treatments. DNA-Free total RNA from different treatments of 2-week-old seedlings were analyzed by semi-quantitative RT-PCR. The actin (GenBank accession no. AB181991) transcript was used an internal control. Control reaction was performed using non-reverse transcribed RNA as a template in order to eliminate the contamination from genomic DNA. No amplification product was detected in control reactions. Experiments were repeated at least four times with similar results. (A) Expression of TaNADP-ME1 in leaf under various treatments. (B) Expression of TaNADP-ME2 in leaf under different various treatments.
These results demonstrate that exogenous hormones (ABA and SA), as well as salt, low temperature and drought stresses can regulate the expressions of TaNADP-ME1 and TaNADP-ME2 in wheat, suggesting that the two NADP-ME genes play an roles in response of wheat to ABA and SA, as well as salt, low temperature and drought stresses.
3.4 TaNADP-ME1 and TaNADP-ME2 are light-responsive genes
To determine whether the two NADP-ME genes are responsive to light in leaf, we studied the expression of the two NADP-ME genes under dark treatment. RNA was extracted from leaf at different periods after transferring seedlings from light into dark condition. Control was performed without treatment and named 0 h. For TaNADP-ME1 (Fig. 4A), the expression amount went down in the first 12 h after treatment, reached the lowest point at 12 h and then began to recover till 48 h. Under the same conditions, TaNADP-ME2 expression was down-regulated and maintained a much lower level from 6 h till to 48 h (Fig. 4B). This suggests that TaNADP-ME1 and TaNADP-ME2 genes are light-responsive genes.
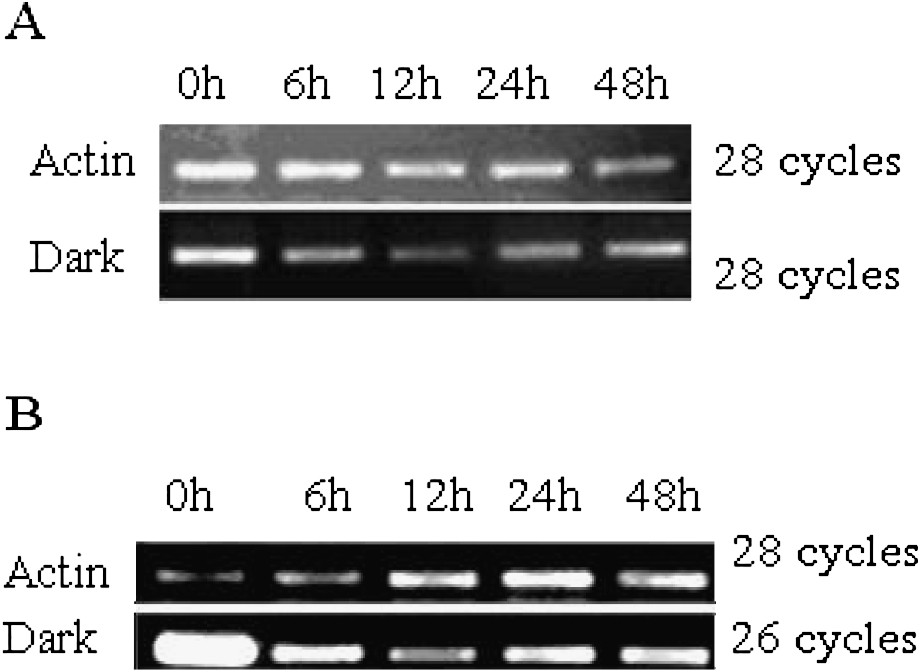
Expression of TaNADP-ME in leaves under dark treatment. (A) Expression of TaNADP-ME1 in leaf under dark treatment. (B) Expression of TaNADP-ME2 in leaves under dark treatment.
3.5 Phylogenetic relationships among NADP-MEs of different plants
To establish the phylogenetic relationships among wheat NADP-MEs and others, an unrooted phylogenetic tree was constructed with various plant full-length NADP-ME amino acid sequences by the neighbor-joining method. Thirty-five plant NADP-MEs were classed into four distinct groups (Fig. 5), forming the resulting phylogenetic tree: Group I consists of dicot cytosolic proteins, Group II consists of monocots, Group III encompasses dicot plastidic NADP-MEs, and Group IV includes dicot and monocot cytosolic proteins.
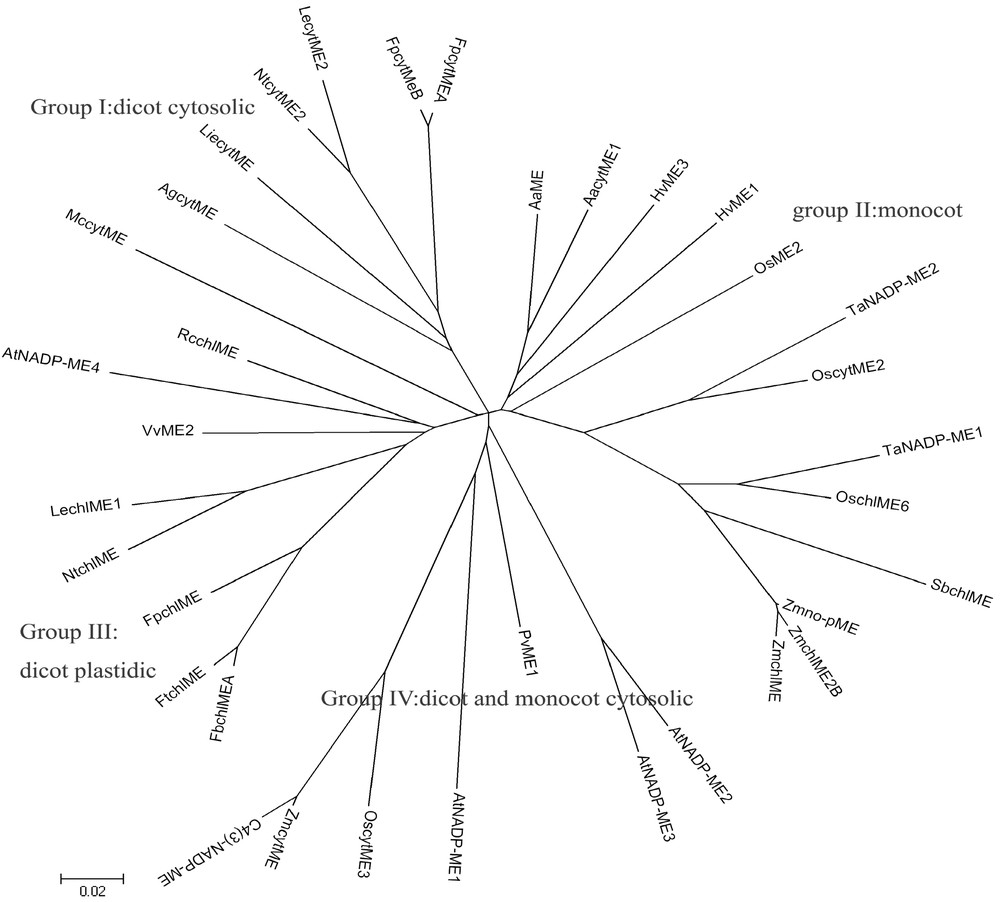
Phylogenetic tree of complete plant NADP-ME. Phylogenetic tree was constructed by the neighbor-joining (NJ) method of mega 4.0 program. Bootstrap analysis was computed with 1000 replicates. The sequences used in this figure, in addition to the two wheat NADP-MEs, are as follows: Zmno-pME (Zea mays non-photosynthetic NADP-malic enzyme, AY315822), ZmchlME2B (Zea mays, AY040616), ZmchlMe (Zea mays, U39958), ZmcytME (Zea mays, AJ224847), C4(3)-NADP-ME (Zea mays, AY864063), OscytME2 (Oryza sativa, AY435404), OscytME3 (Oryza sativa, AY444338), OsME2 (Oryza sativa, AB053295), OschlME6 (Oryza sativa, D16499), AtNADP-ME1 (Arabidopsis thaliana, AY062734), AtNADP-ME2 (Arabidopsis thaliana, AT5G11670), AtNADP-ME3 (Arabidopsis thaliana, AT5G25880), AtNADP-ME4 (Arabidopsis thaliana, AT1G79750), NtcytME2 (Nicotiana tabacum, DQ923118), NtchlME (Nicotiana tabacum, DQ923119), HvME1 (Hydrilla verticillata, AY594687), HvME3 (Hydrilla verticillata, AY594689), SbchlME (Sorghum bicolor, AY274836), PvME1 (Phaseolus vulgaris, X80051), RcchlME (Ricinus communis, AF262997), FpcytMEB (Flaveria pringlei, AF288921), FpcytMEA (Flaveria pringlei, AF288920), FpchlME (Flaveria pringlei, P36444), FbchlMEA (Flaveria bidentis, AY863144), FtchlME (Flaveria trinervia, P22178), AaME (Aloe arborescens, AB005808), AacytME1 (Aloe arborescens, AB016804), LechlME1 (Lycopersicon esculentum, AF001269), LecytME2 (Lycopersicon esculentum, AF001270), LiecytME (Lithospermum erythrorhizon, AB078329), VvME2 (Vitis vinifera, U67426), MccytME (Mesembryanthemum crystallinum, P37223), AgcytME (Apium graveolens, AJ132257).
It appears that TaNADP-ME1 and TaNADP-ME2 belong to Group II and closely resemble OschlME6 and OscytME2, respectively (Fig. 5). In this tree, the dicots were divided into cytosolic and plastidic proteins, which suggest that dicot NADP-MEs may originate from two different ancestral genes. No group was separated in the monocots. MccytME is placed into Group III rather than Group I. ZmcytME, C4(3)-NADP-ME and OscytME3 were placed in Group IV rather than in monocots. The discrepancy of them with the other monocots NADP-MEs indicates there are two independent evolution branches of NADP-ME in monocots. One branch represents the evolution of C4 cytosolic isoforms and the other represents the evolution of plastidic isoforms of C4 plants. In addition, the cooccurrence of plastidic and cytosolic NADP-MEs in monocot (Group II) suggests plastidic NADP-MEs may have arisen from a kind of cytosolic isoform by gene recombination and duplication to get transit peptide sequences (cTP for the chloroplast) during the evolution. This phylogenetic tree displays the complexity of plant NADP-ME evolution. The ambiguities remained may be resolved with further NADP-ME sequences and functional analysis.
4 Discussion
NADP-ME is an ubiquitous enzyme and is involved many metabolic pathways. We isolated and characterized the two NADP-ME genes (TaNADP-ME1 and TaNADP-ME2) from hexaploid wheat. The TaNADP-ME1 gene [NCBI: EU170134] has a full-length of 2392 bp with an open reading frame (ORF) of 1944 bp and encodes 647 amino acids. The complete TaNADP-ME2 gene [NCBI: EU082065] contains 2210 bp, has an open reading frame (ORF) of 1713 bp and encodes 570 amino acids. The similarities of these two genes in whole amino acids are 73.26% and in nucleotide sequences are 64.08%. TaNADP-ME1 and TaNADP-ME2 encode a putative plastidic isoform and a cytosolic isoform, respectively. Five highly conserved sites are found in two NADP-ME genes, while the conserved I228 is substituted by T in Site II of TaNADP-ME1 and T121 is altered to V in Site I of TaNADP-ME2. These alterations in wheat are unique in reported plant NADP-ME and whether these changes might lead to variations in enzyme activity needs further investigation. The multiple sequence alignment shows that TaNADP-ME1 and TaNADP-ME2 is similar to OschlME6 and OscytME2, respectively [48–53].
Tissue-specific analysis showed that TaNADP-ME1 and TaNADP-ME2 are both expressed in roots, stems and leaves of wheat during the light period without treatment. Moreover, TaNADP-ME1 is a leaf-abundant isoform and the transcript accumulations of TaNADP-ME2 in leaves and in roots are almost equal in quantity. Previous researchers have reported tissue-specific expression of plastidic and cytosolic isoforms. To date, at least three NADP-ME genes have been found in maize, namely a plastidic photosynthetic isoform (ZmChlMe1), a plastidic non-photosynthetic isoform (ZmChlMe2) and a cytosolic isoform (ZmcytME). ZmChlMe1 is about 62 kDa, expressed in leaf. ZmChlMe2 is approximately 66 kDa, constitutively expressed with its root-abundance. ZmcytME is expressed in root epidermis [7,28,34]. In rice, at least three cytosolic isoforms and one plastidic isoform have been identified and two cytosolic isoforms have been found to confer salt stress tolerance [8,9]. Transcriptional levels of plastidic isoforms are almost identical in mature tissues, while cytosolic isoforms have different tissue-specific expression, as exemplified by OscytME2 abundance in root [21]. In tobacco, one plastidic and one cytosolic isoform have been reported, and their transcriptional levels are higher in leaves than that in roots despite their constitutive expression in mature tissues [32,43–53]. With respect to Arabidopsis thaliana, there are four NADP-ME genes. AtNADP-ME1 is restricted to roots, AtNADP-ME3 to trichomes and pollen, while AtNADP-ME2 and AtNADP-ME4 are constitutively expressed. What is more, AtNADP-ME2 is an active isoform [19,35]. In view of the number of identified NADP-ME genes in the above-mentioned plants, we can speculate that as a hexaploid with a greater genome, wheat may contain other NADP-ME genes, which have not been isolated yet, in addition to the two genes isolated from our work. These results, together with our work, indicate that the coexpression of plastidic and cytosolic NADP-MEs may be universal for all plant species.
Both genes are classified into Group II as detected by phylogenetic analysis (Group II; Fig. 5). This phylogenetic tree is similar to others previously reported [21,32,35]. One difference with the other analysis is that MccytME is placed into Group III rather than Group I. The reason may be the difference of programs used to construct the phylogenetic tree [Fig. 5].
Semi-quantitative RT-PCR analysis indicate that exogenous hormones (ABA and SA), as well as salt, low temperature and drought stresses regulate the expressions of TaNADP-ME1 and TaNADP-ME2 in wheat, suggesting that the two NADP-ME genes play important roles in response of wheat plant to ABA and SA, as well as salt, low temperature and drought stresses. It is interesting that SA can regulate the expressions of the two NADP-ME genes because there is no other report about the relationship between NADP-ME and SA. This suggests that SA may play an important role such as signal transduction in the NADP-ME response to stress. Previous studies have showed that plastidic and most cytosolic NADP-ME genes were responsive to ABA, PEG, low temperature and NaCl [3,8,9,19,21,32,35]. Likewise, Casati et al. [30] reported that NADP-ME activity in wheat leaf increased when treated with ABA, cellulase and macerozyme. Therefore, we presume that the expression regulation of TaNADP-ME1 and TaNADP-ME2 genes under different abiotic stresses is under post-transcriptional or translational level. Transgenic Arabidopsis plants over-expressing the cytosolic NADP-ME gene of rice enhanced the tolerance of those plants to salt and osmotic stresses [8,9]. But whether these two wheat genes enhance plant tolerance to some kind of abiotic stress remains unknown [43,44]. So the detailed regulation mechanism and function of these two genes under abiotic stresses or exogenous plant hormones need to be comprehensively studied in the future.
Previous researches have reported that several enzymes such as rubisco, glyceraldehyde 3-phosphate dehydrogenase, ribulose 5-phosphate kinase, fructose-1,6-bisphosphatase, PPDK or NADP-MDH and maize chloroplastic NADP-malic enzyme were activated by light [7,27,36–40]. In addition, photosynthetic isoforms of NADP-ME in C4 plants and CAM plants have been reported to range in size from 62 to 67 kDa [25,41]. Furthermore, apart from the character of the light-regulation and subunit size, the photosynthetic MEs can also be distinguished from their non-photosynthetic counterparts by optimum pH, cellular or subcellular localization (chloroplast of mesophyll cells), and a higher affinity for malate [2,6,7,29,48–52]. Although TaNADP-ME1 and TaNADP-ME2 are light-responsive genes by the semi-quantitative RT-PCR analysis under dark treatment, we are not certain if they belong to photosynthetic MEs and take part in photosynthesis. So further biochemical experiments are needed to verify the function of the two genes.
5 Conclusions
Here, we identified two NADP-ME genes from hexaploid wheat (Triticum aestivum L.), named TaNADP-ME1 [NCBI: EU170134] and TaNADP-ME2 [NCBI: EU082065]. Bioinformatics analysis display that TaNADP-ME1 [NCBI: EU170134] encodes a putative plastidic isoform and TaNADP-ME2 [NCBI: EU082065] encodes a cytosolic counterpart. The two NADP-ME isoforms share an identity of 73.26% in whole amino acids and 64.08% in nucleotide sequences. The phylogenetic analysis deciphers the two NADP-MEs as belonging to the monocots (Group II), which closely resemble OschlME6 and OscytME2, respectively. The two genes are co-expressed in root, stem and leaf, and that TaNADP-ME1 is a leaf-abundant isoform. This result further proves the co-expression of plastidic and cytosolic NADP-MEs may be universal for all plant species. In addition, semi-quantitative RT-PCR analysis show that the two NADP-ME transcripts in wheat leaves respond differently to low temperature, salt, dark and drought stresses stimuli and to exogenous abscisic acid (ABA) and salicylic acid (SA). It suggests the two NADP-ME genes play important roles in response of wheat plant to ABA and SA, as well as low temperature, salt, dark and drought stresses. What is more, TaNADP-ME1 and TaNADP-ME2 genes are light-responsive genes by the semi-quantitative RT-PCR analysis under dark treatment. If further biochemical experiments verify these two genes belong to photosynthetic MEs, it will be helpful to improve photosynthetic character of wheat. So this work lays the foundation for understanding the physiological function of NADP-MEs and improvement the tolerance of wheat under abiotic stress. Finally it will contribute to genetic improvement of wheat [43–47].
Acknowledgements
We thank Dr. Zhang Wensheng, Dr. Xujin, Dr. Jijun and assistant researcher Wang Jing in the Center for Agricultural Resources Research, Institute of Genetic and Developmental Biology, Chinese Academy of Sciences for their invaluable assistance. This work is part of a doctoral degree program and was supported by grants from the National 863 Water-saving of Important Item (2006AA100201), the China National Science and Technology Uphold Plan (2006BAD29B02), the International Cooperative Partner Plan of Chinese Academy of Sciences, the Cooperative and Instructive Foundation of State Key Laboratory of Soil Erosion and Dryland Farming on the Loess Plateau (10501-HZ), the Award Foundation of State Key Laboratory of Soil Erosion and Dryland Farming on the Loess Plateau, and the Open Foundation of State Key Laboratory of Soil Erosion and Dryland Farming on the Loess Plateau (10501-194).


