The olive is the most emblematic tree of the Mediterranean basin [1]. Its domestication is considered to have occurred in the Near-East [2] and it spread further from the East to the West of the Mediterranean basin with human migrations [3,4]. The introduction of the olive by the Greeks in Marseille 2500 years ago, is well documented everywhere [3,4], and, although the oleaster and the olive were already present, the Greeks probably introduced new olive cultivation methods. Recent molecular studies have brought information on the oleaster life history since the last Ice Age [5] as well as on cultivar relationships. Several domestication events have probably occurred for this species as demonstrated by the diversity of cultivars based on chlorotypes; thus genetics may recognize them, but the chronology of the events should be based on archaeology remains [6]. Consequently, the olive domestication origin is controversial, and should be revisited as should the hypothesis that the olive has been introduced into the West from the East of the Mediterranean basin. Olive cultivars display huge diversity. The question is whether they differentiate after domestication or if they have several origins.
The olive is the cultivated form, whereas the oleaster is the wild form of Olea europaea subsp. europaea [7]. They are called var. europaea and var. sylvestris, respectively [8]. The crop is propagated either by cuttings or grafts and therefore cultivars are clones. The transition between the oleaster and the olive is based on the size of the pit remains in artefact records. Nevertheless, they are more than 2000 cultivars in the Mediterranean basin that displays huge diversity based on fruit morphology and pit size and morphology and several modern cultivars display as small pits as the oleaster, making the distinction criteria doubtful [9].
Be that as it may, the olive is considered as having been domesticated during the early Neolithic in the Near-East, based on archaeological remains [10]. Several authors underlined the need to cross genetics with archaeology to unravel domestication origins [11]. Traces of olive exploitation have been recorded in the Portugal–Spain Extremadura [12,13], suggesting an early domestication in this region.
Domestication from a wild species to a crop has occurred usually once in the life history of most species [11]. However, for rice and cereals, two or more events have been recorded [14,15]. The crop usually displays a bottleneck of diversity in comparison to the wild due to genetic drift. However, genes under selection are not yet identified for the olive due to a gap in knowledge of the reasons why people have domesticated it. Probably its fruits were used directly, thus leading to increase fruit size. However, we have no evidence whether fruits were used for cakes or oil. It is not obvious that the oleaster was domesticated for its oil. It is logic to think that pickling of the olive wood – since it burns even when wet and was used for feeding animals – has led to domesticate the olive for its fruits [16]. Olive oil uses are numerous and the first uses were probably shamanistic and to burn oil in lamps. The advantage of olive oil is that it burns without smoke; the feed use is documented later, during the Bronze age [17].
Recently, molecular marker studies, both nuclear and cytoplasmic, have revealed that oleasters survived the last Ice Age in eleven refugees [6,18]. Surprisingly, the refugees are less numerous in the East (two in Turkey + Cyprus and Palestine), than in the central Mediterranean (five in Corsica, France; Tunisia) and in the West (four in Morocco, Algeria; Spain, and France). The role Tunisia plays in the central Mediterranean has never been suspected for the oleaster [18]. Nevertheless, the chlorotype genetic structure did not fit with the refugees. The main genetic structure difference shown is based on chloroplast DNA polymorphisms (Fig. 1, A, B) from about 1500 trees [18]. We have found the chlorotype CE1 almost unique in the East (Continent, and Greek Islands), except in Cyprus where it spread with the chlorotype CE2, whereas in the West the major chlorotypes are CE1 COM with two variants (COM1 and COM2) for Spain and the main islands (Sicily, Sardinia, Corsica and the Balearic islands) and the chlorotype CCK mainly in the North of Africa; this suggests that the chlorotype came from ancestral Olea europaea subsp. and were assimilated into the oleaster by introgressive hybridization before the last Ice Age [19]. CE1 is present everywhere. Probably, this genetic structure is the result of gene flow from cultivars displaced by humans and with the wild olive [7,20].
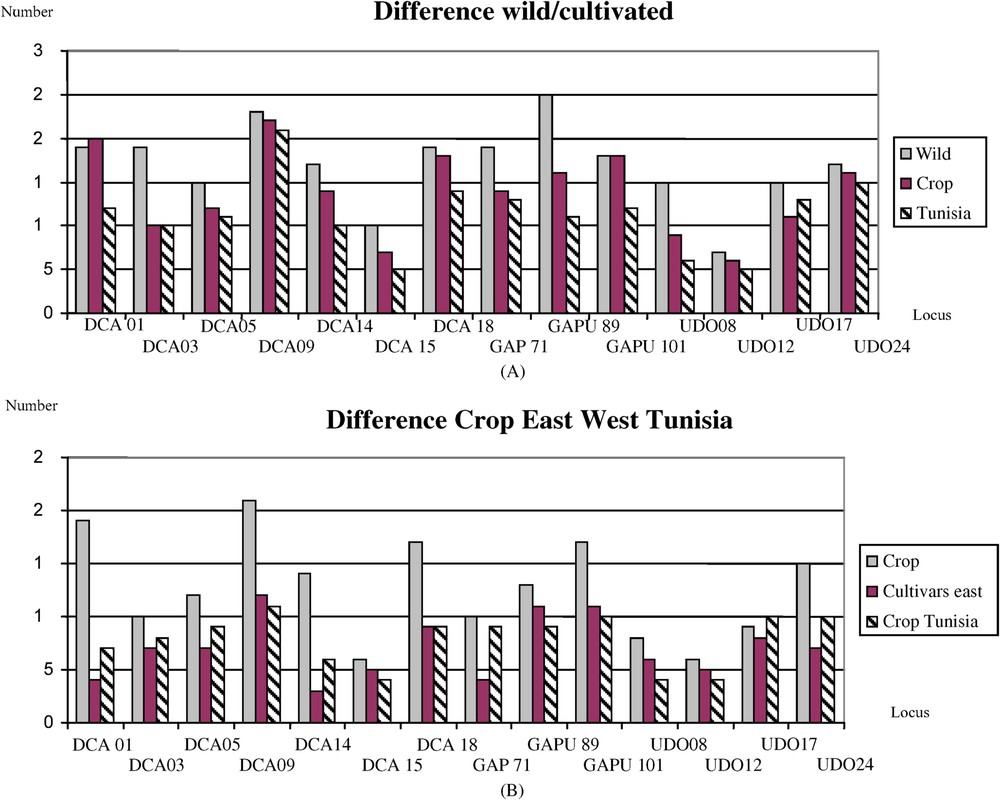
A: Comparison of allele numbers locus a locus between cultivars, oleasters and Tunisian oleasters; B: Comparison of allele numbers for cultivars locus a locus between west and east of the Mediterranean and Tunisian.
A : Comparaison du nombre d'allèles entre cultivars, oléastres et les oléastres de Tunisie ; B : Comparaison du nombre d'allèles par locus entre l'Est, l'ouest de la Méditerranée et la Tunisie.
Gene flow is indicated by the reduced natural spread of the wild olive in comparison to the spread of the cultivars by humans. The spread of the wild is limited to the Mediterranean basin, whereas the cultivars spread in sub-desert regions (South of Tunisia, Morocco, and Libya) and to mountains (Lebanon, Greece, Italy, Spain, France, Portugal) where the oleaster cannot survive alone. Recently, humans have moved the olive tree to all countries with a Mediterranean climate (Africa, Australia, Asia, and America) [1].
Moreover, several approaches have failed to reveal a clear genetic structure in olive cultivars based on different molecular markers, and indeed, olive uses. Based on several types of molecular markers cultivars form a compact genetic group difficult to split based on olive uses, fruit morphology, cultivar locations and on their local names [19]. Typically, dendrogram mixing olive and oleaster trees display one branch that clusters most of the cultivars and oleasters from the East [21,22]. The other branch clusters most oleaster trees from the West and a few cultivars from Corsica, Algeria, Tunisia and some landraces. This suggests that most cultivars have tight relationships with eastern oleasters, whereas the cultivars from the second branch have kinship relationships with the western oleasters. In the frame of the hypothesis that several domestication events have occurred we expected that cultivars should have ancestors in at least one or more ancestral oleaster lineage. Moreover, due to gene flow we also expected that crosses may have occurred between different origins. We also expected a weak bottleneck of diversity between the olive and the oleasters. Probably, finally, some ancestral oleaster population may have not contributed to cultivars.
To check these expectations we have sampled a large set of 411 cultivars from all Mediterranean countries and 958 oleaster trees from seventy populations that thrive naturally all around the Mediterranean basin, to geographically anchor olive origin in oleaster forests. All trees were genotyped with 16 microsatellite markers 14 for the nuclear and 2 for the chloroplast DNA to benefit from the strong geographic structure in oleaster [6,23,24].
Overall, the 958 oleaster display 217 alleles, thus an equivalent allelic diversity to the 411 olive cultivars that displays 194 alleles. We also computed for the East, the West and Tunisian stock and found that the three regions display equivalent allelic frequencies at each locus. No allele can identify genuine oleaster versus olive cultivar. However, due to different sample size between oleasters and cultivars we weighted the differences and we concluded that the bottleneck in diversity is not meaningful (Fig. 2).

Comparison of allele numbers locus a locus between cultivars and oleasters.
Comparaison du nombre d'allèles à chaque locus entre cultivars et oléastres.
We also used Bayesian clustering on cultivars and found that about half of the cultivar trees were assigned to nine genetic lineages, whereas the other admixed in two or more lineages. We found that cultivars admixed to nine of the oleaster lineages spreading in nine geographic areas. This indicates that those olive cultivars share tight kinship relationships with most of the oleaster genetic lineages. The question is therefore whether the nine genetic clusters may be considered as nine origins of domestication.
Obviously, the present name of a cultivar cannot warrant its geographic origin. This results from cultivar displacements all around the Mediterranean basin and they received new local names (Fig. 3). We treated our data with classical methods (genetic distances, aggregation of distances, factorial correspondence analyses). Our data sustain domestication centres in Turkey and Palestine in the East. Such a centre in Palestine has already been revealed [2], whereas a centre in Turkey was never suspected. Some cultivars carrying presently French, Italian or Spanish names have ancestors in the Near East (Turkey, Palestine, and Cyprus) or in North Africa (Tunisia and Algeria); most of them carry the CE1 chlorotype or CCK or COM [5]. In Cyprus, we found oleasters carrying the chlorotype CE2, and consequently cultivars carrying this chlorotype should derive from those of Cyprus, but a modern cultivar carrying CE2 may belong randomly to another lineage after several backcrosses, an example is ‘Galega’ from Portugal.

Each camembert represents the proportion for a cultivar of the genome from the different GRPs (see caption upper right). Chloroplast DNA contribution is indicated by the smaller circle (see caption bottom left).
Chaque camembert pour un cultivar représente la contribution dans son génome de chacune des GRPs (voir la légende en haut à droite). La contribution de l'ADN du chloroplaste est indiquée par le petit cercle (voir la légende en bas à gauche).
Several domestication centres were revealed in the West: in North Africa: Tunisia, Algeria, Morocco (three centres), in Corsica (one centre), in Spain (three centres), and in France (one centre). North Africa area displays oleasters that carry either chlorotypes CCK, COM, COM1, COM2 [24]; thus cultivars derived maternally from those oleasters carry the same chlorotype, but here our results show that cultivars not carrying these chlorotypes may also admixed to this centre. The cultivars that display the chlorotype COM, COM1, or COM2 from central Mediterranean revealed that they derived maternally from oleasters of this area. Archaeological data sustain one domestication centre in Portuguese Estremadura [11,25], and we suspected that some cultivars from Spain (such as ‘Picual’) which display the chlorotype COM, have a local origin in local oleasters.
One cultivar from Corsica (‘Sabina’) has all morphological characteristics of an oleaster displaying the chlorotype COM, and may correspond to a local domestication event as supported by archaeological evidence [26,27]. Archaeological data in other regions are scarce and if abundant, such as olive pits in Palestine, their genetic information has not yet been compared to modern olive cultivars [28].
We concluded that olive cultivars have nine origins and about half of cultivars result from crosses between one, two or more lineages. Three lineages are sustained in the East, and in the west (Spain) in Extremadura and in Corsica France, based on archaeological evidence as corresponding to olive domestication centres. Tunisia is a pole of diversity for the oleaster, cumulating introduction of cultivars from the East by Phoenicians plus local domestication from oleaster [29]. Moreover, important gene flow between cultivars and wild olive, in both directions, still pose questions on historic relations (migration) and genetic (selection) of the olive.


