1 Introduction
In wheat kernels, starch is the primary sink product, and accounts for approximately 65 to 75% of seed weight. ADP-glucose pyrophosphorylase (AGPase) carries out the first committed step in the starch synthesis, and increasing AGPase activities significantly improves seed weights [1–3]. AGPase in plants is a heterotetrameric enzyme composed of two large subunits (LSU) and two small subunits (SSU) [4,5]. Both subunits are essential for normal AGPase activity. SSUs are capable of forming a homotetrameric enzyme exhibiting normal catalytic properties but are defective in allosteric regulatory properties. LSUs are incapable of forming an active enzyme, and increase the allosteric regulatory response of SSUs to effectors (3-PGA or Pi) [6].
There are two distinct AGPases corresponding to cytosolic and plastidial isoforms [7–9]. Accordingly, there are four types of AGPase subunits in plant cells: cytosolic SSU, plastidial SSU, cytosolic LSU and plastidial LSU [10]. In most plant cells, AGPase occurs exclusively in the plastids. However, in the endosperms of the Poaceae (grass family), including the economically important cereal crop plants maize (Zea mays), rice (Oryza sativa), barley (Hordeum vulgare), and wheat (Triticum aestivum), AGPase is mainly present in the cytosol. In these tissues, ADP-glucose synthesized by the cytosolic isoform of AGPase is imported into the plastids for starch synthesis via an ADP-glucose transporter [11].
In barley, two genes encoding the SSUs (Hv.AGP.S.1 and Hv.AGP.S.2) of AGPase were identified. Hv.AGP.S.2 [NCBI: AF537363] gave rise to a single transcript encoding the plastidial SSU [12]. It could be responsible for most or all of the plastidial SSU in a range of nonphotosynthetic plant organs including the embryo, and probably made some contribution to SSU of AGPase in leaves. However, Hv.AGP.S.1 gave rises to two transcripts by use of the alternative first exons in kernels [8]. One of the alternative first exons, exon1b (Hv.AGP.S.1b) [NCBI: Z48563], encoded a transit peptide for targeting its precursor protein to plastids. The other first exon, exon 1a (Hv.AGP.S.1a) [NCBI: Z48562], was shorter than exon 1b and did not include a predicted transit peptide. This suggested that one of the proteins encoded by Hv.AGP.S.1 was plastidial (Hv.AGP.S.1b) and the other was cytosolic (Hv.AGP.S.1a).
Burton et al. [10] isolated 5′ terminal cDNA sequences of Ta.AGP.S.1a (260 bp) and Ta.AGP.S.1b (353 bp) in kernels of a non-Chinese wheat cultivar (Bobwhite), and sequence analysis indicated that they were also derived from the Ta.AGP.S.1 gene [NCBI: AF536819] by use of the alternative first exons. In this study, a novel Ta.AGP.S.1b transcript [NCBI: EU586278] was isolated from Chinese wheat cultivars. Compared with the reported Hv.AGP.S1b [NCBI: Z48563], EU586278 manuscript was characterized with a long lacked fragment at its 5′ terminal, resulting in a shorten transit peptide. Partial genomic DNA sequence corresponding to 5′ terminal of EU576278 transcript was also amplified to explore the possible mechanism on the lacked fragment. Also, the chromosome location and expression pattern of EU586278 in different organs were also further explored.
2 Materials and methods
2.1 Total RNA extraction and cDNA synthesis
The developing endosperms at the 15th day after anthesis for 22 Chinese common wheat cultivars, Yujiao 2, Yanzhan 4110, Yunong 949, Yumai 34, Zhengmai 9023, Yunong 202, Xinmai 9, Aikang 58, Luomai 21, Zhoumai 18, Zhoumai 17, Yumai 49, Lumai 22, Lankao Aizao 8, Yannong 15, Shanyou 225, Xiaoyan 81, Kaimai 18, Neixiang 188, Luomai 4, Jimai 20, Yumai 18 and two non-Chinese wheat cultivars (Bobwhite and Chinese Spring) were dissected and used for RNA extraction.
Total RNA was extracted using Trizol reagents (Invitrogen) and its integrity was confirmed by gel electrophoresis. cDNA was prepared from 2 μg of total RNA with Thermoscript RT (Toyobo) and an oligod(T)18 primer at a temperature of 42°C according to the manufacturer's instructions.
2.2 Cloning of Ta.AGP.S1a and Ta.AGP.S1b transcripts in kernels of Chinese wheat cultivars
Based on known nucleotide sequence of Hv.AGP.S1b transcript [NCBI: Z48563] in barley, the cDNA sequences of Ta.AGP.S1bs from two wheat cultivars (Bobwhite and Yujiao 2) were amplified with a pair of primers: 5′-CCACCTCAATGGCGATGGC-3′ (forward) and 5′-CCAGGGGCACTTCGCGTAA-3′ (reverse) (at the positions from 9 nt to 1749 nt in Z48563) (P1 primers). Cycle parameters were 95°C for 3 min, 35 cycles of 95°C for 50 s, 55°C for 50 s, 72°C for 2 min 30 s, and an extension of 72°C for 8 min.
5′ terminal cDNA sequences of Ta.AGP.S1b transcripts in the above 22 Chinese wheat cultivars and two non-Chinese wheat cultivars (Bobwhite and Chinese Spring) were also amplified with a pair of primers: 5′-GCCTCCCCCTCCAAGAT-3’ (forward) and 5′-TGGAGTCTGACACGGCAC-3′ (reverse) (at the positions from 35 nt to 221 nt in Z48563) (P2 primers). Cycle parameters were 95°C for 5 min, 30 cycles of 95°C for 30 s, 53°C for 30 s, 72°C for 1 min, and an extension of 72°C for 8 min.
cDNA sequence of Ta.AGP.S1a transcript in Yujiao two cultivar was amplified using the primers: 5′-AATGGATGTGCCTTTGGC-3′ (forward) and 5′-ATGCAAGTAGCTCTGTCC-3′ (reverse) (at the positions from 16 nt to 1480 nt in X66080) (P3 primers). Cycle parameters were 95°C for 8 min, 30 cycles of 95°C for 40 s, 56°C for 40 s, 72°C for 2 min, and an extension of 72°C for 8 min.
The above amplified fragments were isolated from gels, purified using gel purification kit (Promega), and cloned into pMD20-T vector (Takara). Each product was completely sequenced three times using the Applied Biosystems 3710 DNA capillary sequence.
2.3 The putative transit peptides analysis and phylogenetic tree construction for plant AGP.S1bs
TargetP version 1.1 was used to analyze the transit peptides of AGP.S1bs in wheat and other crops without cutoff, including the cleavage site prediction (http://www.cbs.dtu.dk/services/TargetP/) [13]. Amino acid sequence analysis on transit peptides of plant AGP.S1bs was done using DNAMAN 3.0.
2.4 Cloning of 5′ partial genomic DNA sequence of Ta.AGP.S.1 gene in Yujiao 2 wheat cultivar
Genomic DNA was isolated in Yujiao 2 cultivar using the SDS method. Based on the reported Ta.AGP.S.1 gene [NCBI: AF536819], a pair of primers were designed to amplify the DNA sequence of Ta.AGP.S.1 gene corresponding to the lacked fragment of EU586278 transcript. The primers were: 5′-TAGTAGCCCGTCAAGTAGCA-3′ (forward) and 5′-CTCCACCACATCCGAAAT-3′ (reverse) (at the positions from 680 nt to 1813 nt in AF536819) (P4 primers). Cycle parameters were 95°C for 8 min, 30 cycles of 95°C for 40 s, 53°C for 40 s, 72°C for 1 min 30 s, and an extension of 72°C for 10 min. The amplified 1124 bp fragment was also purified, linked to pMD20-T vector, and sequenced three times.
2.5 Chromosomal location of Ta.AGP.S.1 gene
After searching the Ta.AGP.S.1 sequence against the expressed sequence tags (ESTs) in GrainGenes (http://wheat.pw.usda.gov), the chromosomal location and copy number of Ta.AGP.S1b transcripts were determined.
2.6 Transcription levels for EU586278 transcript in different organs of Yujiao 2 cultivar
Semi-quantitative RT-PCR was used to measure transcription levels of EU586278 in different organs of Yujiao 2 cultivar. Total RNAs of roots, stems, leaves and endosperms (embryos were dissected from kernels) at the 15th day after anthesis were used to analyze the tissue-special expression of EU586278 with P1 primers. Each RNA sample was evaluated by β-actin gene using a pair of primers 5′-GTTCCAATCTATGAGGGATACACGC-3′ (forward) and 5′-GAACCTCCACTGAGAACAACATTACC 3′ (reverse) that amplified a 422 bp cDNA fragment.
3 Results
3.1 Characterization on cDNA sequences for Ta.AGP.S1b transcripts in Chinese common wheat cultivars
In this study, P1 primers were designed to amplify partial cDNA sequences of Ta.AGP.S1b transcripts in the kernels of a common Chinese wheat cultivar (Yujiao 2) and a non-Chinese wheat cultivar (Bobwhite), respectively. The electrophoretic results indicated that amplified fragment from Bobwhite was slightly longer than that from Yujiao 2 (Fig. 1). The sequencing results further showed that the amplified Ta.AGP.S1b in Bobwhite was 1748 bp [NCBI: FJ643609], but 1631 bp [NCBI: EU586278] in Yujiao 2 (Fig. 2).
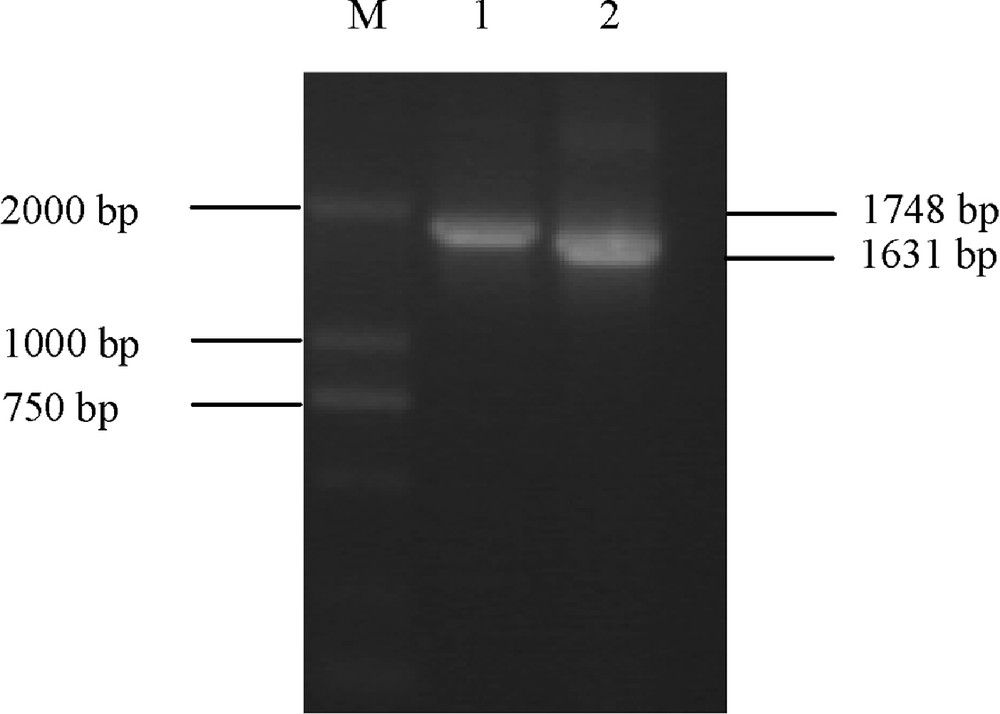
The amplified cDNA sequences for Ta.AGP.S1b transcripts in both Bobwhite (lane 1) and Yujiao (lane 2) cultivars, respectively. M: molecular mass standard.

Comparison of cDNA sequences encoding AGP.S1b transcripts in wheat and barley. EU586278 and FJ643609, cDNA sequences of Ta.AGP.S1b transcripts from Yujiao 2 and Bobwhite wheat cultivars, respectively; Z48563, cDNA sequence of Hv.AGP.S1b [8]. AF536819C, partial 5′ cDNA sequence of Ta.AGP.S1b previously isolated in Bobwhite cultivar [10]. cDNA sequences encoding the putative transit peptides of the above surveyed AGP.S1bs using TargetP software (Fig. 4) were underlined. Identical bases were shown in white on black. P1 and P2 primers were used to amplify CDS and 5′terminal regions of Ta.AGP.S1b in Yujiao 2 wheat cultivar, respectively. EF405961, Ta.AGP.S1a isolated in Yujiao 2, and its primers (P3) were not shown. The transcript start codon (ATG) and end codon (TGA) were shown in asterisks. P1-F and P1-R, P2-F and P2-R, P1 and P2 forward and reverse primers, respectively.
FJ643609 shared 97.14% identity to Hv.AGP.S1b [NCBI: Z48563]. However, EU586278 shared only 92.39% and 90.89% identities to those of FJ643609 and Z48563, respectively. Its lower identities to Z48563 and FJ643609 were mainly because of a long lacked fragment (114 bp at the positions from 68 nt to 181 nt in Z48563, or 117 bp at the positions from 60 nt to 176 nt in FJ643609, respectively) at its 5′ terminal (Fig. 2).
To explore whether there was the above lacked fragment of EU586278 in other Chinese wheat cultivars, another pair of primers (P2) were used to amplify the 5′ terminal of Ta.AGP.S1b transcripts. The results indicated that 190 bp fragments were amplified in two non-Chinese wheat cultivars (Bobwhite and Chinese Spring), but 73 bp fragments in all 22 surveyed Chinese common wheat cultivars (Fig. 3), indicating that the lacked 117 bp cDNA sequences at 5′ terminal of Ta.AGP.S1bs could be universally present in Chinese common wheat cultivars.
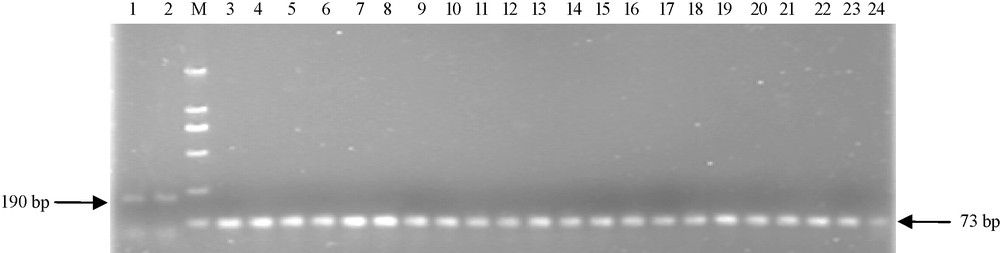
The 5′ terminal sequences for Ta.AGP.S1bs from the kernels of both 22 Chinese and two non-Chinese wheat cultivars. M, Molecular mass standard; lanes 1 and 2, 5′ terminal sequences of Ta.AGP.S1bs from two non-Chinese wheat cultivars, Bobwhite and Chinese Spring, respectively; lanes 3–24, 5′ terminal sequences of Ta.AGP.S1bs from 22 common Chinese wheat cultivars, Yujiao 2, Yanzhan 4110, Yunong 949, Yumai 34, Zhengmai 9023, Yunong 202, Xinmai 9, Aikang 58, Luomai 21, Zhoumai 18, Zhoumai 17, Yumai 49, Lumai 22, Lankao Aizao 8, Yannong 15, Shanyou 225, Xiaoyan 81, Kaimai 18, Neixiang 188, Luomai 4, Jimai 20 and Yumai 18, respectively. Sequences of P2 primers for this amplification and the predicated fragments from the above wheat cultivars were indicated in Fig. 2.
The amplified cDNA sequence of Ta.AGP.S1a transcript in Yujiao 2 cultivar was 1465 bp [NCBI: EF405961] (Fig. 2), and it shared 99%–98% identities to other Ta.AGP.S1as [NCBI: AF244997, AF492644 and X66080] and 96% identity to Hv.AGP.S1a [NCBI: Z48562] (data not shown). The middle and 3′ regions of EF405961 and EU586278 shared 98% identity, but their 5′ terminal regions were considerably different (Fig. 2). This implied that, as Hv.AGP.S1b [NCBI: Z48563] and Hv.AGP.S1a [NCBI: Z48562] in barley, both EU586278 and EF405961 transcripts in kernels of Chinese common wheat cultivars were also encoded by the same gene (Ta.AGP.S.1) with the different alternative first exons.
CDS (coding sequence) of FJ643609 was 1545 bp, corresponding to a putative protein with 515 amino acid residues. Because of the long lacked fragment, however, CDS of EU586278 contained only 1428 bp, corresponding to a putative protein with 476 amino acid residues (http://www.expasy.org/cgibin/protparam) (Fig. 2). And the calculated molecular mass of EU586278 transcript was only 55.23 kDa, also fewer than that of FJ643609 (59.27 kDa).
3.2 Characterizations on the transit peptides of Ta.AGP.S1bs in Chinese common wheat cultivars
TargetP software (www.cbs.dtu.dk/services/TargetP) is a good tool to predict the transit peptides and their cleavage cites for the proteins which are located in plastids, and has broadly been used as far [13–15]. Using TargetP software, lengths of most predicated transit peptides for plastidial SSUs were 54 to 70 amino acid residues in wheat [NCBI: FJ643609 and AF536819], barley [NCBI: Z48563], maize [NCBI: AF334960], rice [NCBI: AK103906], potato [NCBI: DQ207843], Arabidopsis [NCBI: AY096379], sugar beet [NCBI: X78899], bean [NCBI: X76941], tomato [NCBI: L41126] and rape [NCBI: AJ271162] (Fig. 4 and Table 1). The predicated cleavage sites of all surveyed cereal AGP.S1b transit peptides were the same (RǀA), and amino acid sequences after the cleavage sites were completely identical (Fig. 4). This indicated that, after these SSU precursor proteins were directed into plastids by the transit peptides and their transit peptides were cleaved off by the processing peptidase, the functional amino acid sequences of their matured plastidial SSUs were completely identical.

Comparisons on putative amino acid sequences of.AGP.S1bs among some plants. EU586278, amino acid sequence of Ta.AGP.S1b from Yujiao 2 cultivar; FJ643609, amino acid sequence of Ta.AGP.S1b from Bobwhite; AF536819 P, N terminal sequence of Ta.AGP.S1b encoded by AF536819 gene from Bobwhite; Z48563, amino acid sequence of AGP.S1bs from barley; AF334906, amino acid sequence of AGP.S1b from maize; AK103906, amino acid sequence of AGP.S1b from rice; DQ207843, amino acid sequence of AGP.S1b from potato; AY096379, amino acid sequence of AGP.S1b from Arabidopsis; X78899, amino acid sequence of AGP.S1b from sugar beet; X76941, amino acid sequence of AGP.S1b from bean; L41126, amino acid sequence of AGP.S1b from tomato; AJ271162, amino acid sequence of AGP.S1b from rape. There were high identity of the middle and C terminal sequences of AGP.S1bs among the surveyed crops, and most of identical sequences at their 3′ terminal and middle regions were not displayed. ǀ, the transit peptide cleavage sites were predicated with TargetP software. Identical amino acids were shown in white on black.
Prediction on AGP.S.1bs in higher plants with TargetP software.
| Name | Len | cTP | mTP | SP | Other | Loc | RC | TPlen |
| EU586278 | 475 | 0.727 | 0.090 | 0.074 | 0.145 | C | 3 | 25 |
| Z48563 | 513 | 0.931 | 0.028 | 0.013 | 0.036 | C | 1 | 63 |
| FJ643609 | 514 | 0.943 | 0.019 | 0.009 | 0.036 | C | 1 | 64 |
| AF536819 P | 124 | 0.956 | 0.019 | 0.008 | 0.047 | C | 1 | 62 |
| AF334960 | 517 | 0.976 | 0.062 | 0.001 | 0.040 | C | 1 | 67 |
| AK103906 | 514 | 0.892 | 0.020 | 0.021 | 0.021 | C | 1 | 64 |
| DQ207843 | 521 | 0.987 | 0.014 | 0.003 | 0.071 | C | 1 | 58 |
| AY096379 | 520 | 0.976 | 0.013 | 0.008 | 0.044 | C | 1 | 70 |
| X78899 | 489 | 0.769 | 0.066 | 0.022 | 0.144 | C | 2 | 54 |
| X76941 | 512 | 0.968 | 0.053 | 0.005 | 0.033 | C | 1 | 65 |
| L41126 | 521 | 0.983 | 0.018 | 0.002 | 0.056 | C | 1 | 58 |
| AJ271162 | 520 | 0.955 | 0.082 | 0.003 | 0.025 | C | 1 | 57 |
| cutoff | 0.000 | 0.000 | 0.000 | 0.000 |
Because of the lacked 117 bp fragment, the predicated transit peptides of EU586278 had only 25 amino acid residues, and at least 39 amino acids fewer than those of the above other cereal AGP.S1bs (Fig. 4 and Table 1). However, the transit peptide cleavage site of EU586278 was also RǀA, and its amino acid sequence after the transit peptide was also completely identical to those of the above other cereal AGP.S1bs. These results suggested that the function of the matured Ta.AGP.S1b subunit encoded by EU586278 could not be changed by the lacked fragment.
Length of the transit peptide for EU586278 was only 35.7 to 46.3% of other surveyed plant plastidial SSUs. Because the differences in amino acid sequences of all surveyed plant plastidial SSUs focused on their transit peptides located at N terminal (Fig. 4), a phylogenetic tree was generated from the transit peptide sequences of some cereal plastidial SSUs (Fig. 5). The transit peptide sequence of EU586278 was not clustered together with those of Z48563, AF536819 and FJ643609. These inferred that EU586278 could be a novel Ta.AGP.S1b transcript in wheat. However, the score of reliability coefficient (RC = 3) on the predicated transit peptide cleavage site of EU586278 was lower than those of the above other plant plastidial SSUs (RC = 1) (Table 1).
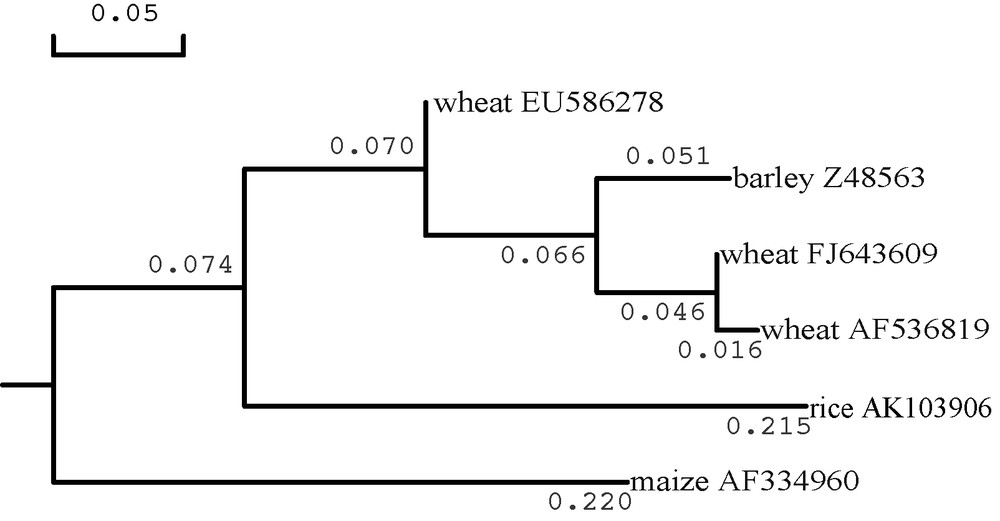
Phylogenetic tree on putative amino acids of the transit peptides from the surveyed AGP.S1bs in cereal plants in DNAMAN3.0. GenBank database accession numbers of the displayed AGP.S1bs were as follows: Triticum aestivum, EU586278, FJ643609 and AF536819; Hordeum vulgare, Z58563; Oryza sativa, AK103906; Zea mays, AF334906.
There are many types of plastids in plant cells, such as chloroplast, amyloid, etc. Using TargetP software, predication further indicated that all surveyed plastidial SSUs, including EU586278, were located in chloroplasts (Table 1), indicating that they could play important roles in transient starch synthesis in leaf.
3.3 Isolation of 5′ partial genomic DNA sequence of Ta.AGP.S.1 gene in Chinese common wheat cultivars
5′ partial DNA sequences of Ta.AGP.S.1 gene (1146 bp, NCBI: FJ907395) were further isolated in Yujiao 2 wheat cultivar. Sequencing indicated that FJ907395 shared 94.26% identity to AF536819, a partial DNA sequence of another Ta.AGP.S.1 gene isolated from Bobwhite cultivar [10]. Moreover, FJ907395 also contained the corresponding lacked cDNA fragment of EU586278 transcript. The lacked fragment of EU586278 was located at the position from 412 nt to 528 nt in FJ907395, and was an uninterrupted fragment (Fig. 6, underlined).

Comparison on partial genomic DNA sequences of Ta.AGP.S.1 genes between a non-Chinese common wheat cultivar (Bobwhite) (AF536819) and a Chinese common wheat cultivar (Yujiao 2) (FJ907395), respectively. Identical nucleotide bases were shown in white on black. Simple repeat sequences in FJ907395 were indicated with lowercases. The lacked cDNA fragment in EU586278 corresponding to FJ643609 was underlined.
There were some simple sequence repeats (gcgcgc, tctctc, etc) in FJ907395, which were located at the positions from 137 nt to 195 nt, 197 nt to 229 nt and 470 nt to 533 nt, respectively. Compared with AF536819, FJ907395 contained many different nucleotide bases (Fig. 6). The especial sequence in FJ907395 corresponding to the lacked cDNA fragment of EU586278 also contained some simple repeat sequences (gcgc, etc), two mutations at the positions of 439 nt (T/C) and 457 nt (C/T), a deletion at the position of 516 nt (C) and an addition at the position of 519 nt (C).
3.4 Chromosomal location of Ta.AGP.S.1 gene
After searching in the 7104 mapped EST sequences in GrainGenes, an EST sequence [NCBI: BE590582], which was 99% identical to both EU586278 and FJ643609, was obtained. This EST was located on wheat chromosome 7AS, 7BS and 7DS. And there were 2 loci in 7AS and 7BS, respectively, but one loci in 7DS. This indicated that two Ta.AGP.S.1 genes, which encoded EU586278 transcript in Chinese common wheat cultivars and FJ643609 transcript in non-Chinese common wheat cultivars, respectively, were located on the same chromosomes (7AS, 7BS and 7DS).
3.5 Tissue-specific expression pattern of EU586278 in Yujiao 2
At 15 d after anthesis, when starch was quickly synthesized in kernels of wheat, the tissue-specific expression pattern of EU586278 was investigated with semi-quantitative RT-PCR in Yujiao 2 cultivar. The transcript products corresponding to EU586278 were abundantly expressed in leaf, moderately in stem and endosperm, but weakly in root (Fig. 7).
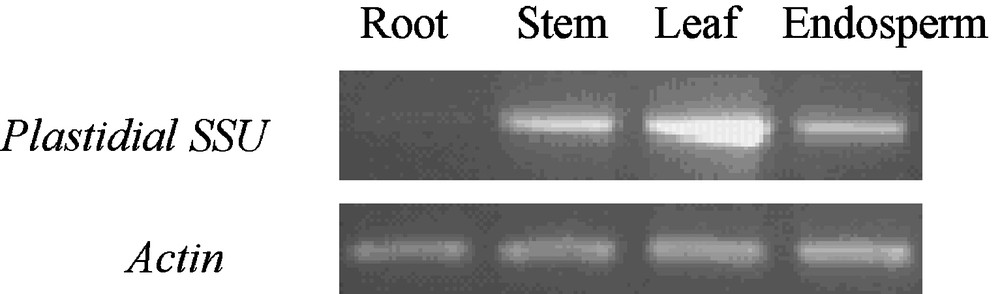
Transcript levels for EU586278 in different organs of Yujiao 2 cultivar.
4 Discussion
In this study, a cDNA sequence of Ta.AGP.S1b transcript [NCBI: EU586278] was isolated from Yujiao 2, a Chinese common wheat cultivar (Figs. 1 and 2). Compared with two homologous transcripts [NCBI: Z48563 and FJ643609], EU586278 lacked a long fragment (117 bp) at its 5′ terminal. Another cDNA sequence [NCBI: EF405961] of Ta.AGP.S1a was also amplified from the same wheat cultivar. Sequence analysis indicated that their 3′ terminal and middle regions of both EF405961 and EU586278 sequences were highly identical, but their 5′ terminal regions clearly different (Fig. 2). This suggested that, as Hv.AGP.S1a [NCBI: Z48562] and Hv.AGP.S1b [NCBI: Z48563] transcripts in barley, both EF405961 (Ta.AGP.S1a) and EU586278 (Ta.AGP.S1b) transcripts in Chinese common wheat cultivars were also encoded by the same gene (Ta. AGP.S.1) with the alternative first exons (Fig. 2), though there was a long missing fragment at the 5′ terminal region of EU586278 transcript. It was reported that, in China's wheat breeding history, a very limited number of wheat varieties were used in the breeding programs, and most of modern Chinese wheat cultivars, including the surveyed wheat in this study, had similar genetic backgrounds [16]. However, there were many differences in genetic backgrounds between modern Chinese common wheat cultivars and non-Chinese common wheat cultivars (Bobwhite, Chinese Spring, etc) [17,18]. Thus, it was speculated that the 117 bp lacked fragments in EU586278, which were widely found in 22 surveyed Chinese common wheat cultivars (Fig. 3), could be also present in most of other modern Chinese wheat cultivars, not in non-Chinese common wheat cultivars.
The 5′ partial genomic DNA sequence [NCBI: FJ907395] of Ta.AGP.S.1 gene, which was corresponded to 5′ terminal of EU586278 transcript, was further isolated from Yujiao 2 cultivar. To be surprised, FJ907395 also contained the corresponding lacked fragment of EU586278 transcript, suggesting the lacked fragment in EU586278 transcript was not present in the genome, but probably occurred at transcription level. Although both EU586278 and FJ643609 transcripts were located at the same loci at chromosomes 7AS, 7BS and 7DS, their corresponding genomic sequences of Ta.AGP.S1 genes (FJ907395 and AF536819, respectively) had many sequence differences (base variations and repeat sequences) at their 5′ terminal regions (Fig. 6). This inferred that these genomic sequence differences between FJ907395 and AF536819 could result in the 5′ terminal lacked fragment of EU586278 transcript when its transcription occurred.
Amino acids of plastidial SSUs in plants shared high identities at their C terminal and middle regions, but clearly different at their N terminal regions which encoded the transit peptides (Figs. 2 and 4). Using TargetP software, the predicated transit peptide of EU586278 contained merely 25 amino acid residues, considerably shorter than those of other surveyed plant plastidial SSUs (57–70 amino acid residues). The differences in amino acid sequences of all surveyed plant plastidial SSUs focused on their transit peptides of their N terminal regions (Fig. 4). Phylogenetic tree analysis indicated that the transit peptide sequence of EU586278 was not also clustered together with those of Z48563, AF536819 and FJ643609 (Fig. 5), suggesting that EU586278 could encode a novel Ta.AGP.S1b transcript.
Although a long lacked fragment at its 5′ terminal, the predicated transit cleavage site (R|A) of EU586278 was as same as those of other cereal AGP.S1bs. And their amino acid residues behind the transit peptides also shared highly identical (Fig. 4). This suggested that the catalytic function of the matured Ta.AGP.S1b subunit encoded by EU586278 could not be affected after its precursor proteins were targeted into plastids and its transit peptides were removed by intraorganellar proteases. EU586278 was located in chloroplasts (Table 1), and it was abundantly expressed in leaf, moderately in stem and endosperm, but weakly in root (Fig. 7). These results were good agreement with previous measurements in barley [8,11] and a non-Chinese common wheat cultivar [10]. This further inferred that function of EU586278 transcript could not be changed by the deletion of a 117 bp fragment.
The score of RC was a measure of how confident TargetP. The RC ranges from 1 (very reliable predication; virtually no false positives detected in the TargetP test set) to 5 (not reliable prediction; many false positives detected) [13]. In this paper, the reliable predication on the transit peptide of EU586278 (RC = 3) was lower than those of other surveyed plant plastidial SSUs (RC = 1–2) (Table 1), inferring that this predication on the transit peptide of EU586278 transcript needed to be confirmed with further study.
Acknowledgements
We thank Prof. Olof Emanuelsson and Prof. Gunnar von Heijne (Sweden Stockholm University) for assistance with TargetP software. We also thank the anonymous reviewers for their valuable comments and in particular their suggestions for further studies. This study was financially supported by National Science Foundation of China (30871472), the State Key Laboratory of Crop Science of China (2009KF01) and the Key Item of Science and Technology of Henan Province (08210240020).


