1 Introduction
The genus Jatropha L. (Euphorbiaceae) includes a large number of species that are distributed all over the world inhabitats ranging from the tropical to the temperate zones. Jatropha curcas is a multipurpose shrub with significant economic importance having the capabilities to rehabilitate the degraded lands [1]. Since the oil crisis of the 1970s and recognition of the limitations of world oil resources, most of the oil importing countries, including India, have been motivated to develop alternative sources of energy. J. curcas has been found to be a highly promising species which can yield oilseed as a source of energy in the form of biodiesel owing to its short gestation period, easy adaptation to different kinds of marginal and semimarginal lands, drought endurance and avoidance by animals. Mutation breeding in tree crops is not considered attractive because of lacunae in conventional breeding like time-consuming, unpredictable results, long juvenile phase, high heterozygosity and fear in loss of the unique genotype. However, studies on induced mutation in fruit crops have been performed particularly in apple, pear, peach, grapevine etc. Mutation studies undertaken at the National Botanical Research Institute (NBRI), Lucknow, India have led to induction of cotyledonary variabilities in J. curcas [2]. Mutation breeding studies in J. curcas have been carried out in Thailand using fast neutrons and have isolated dwarf and early flowering mutants from the M3 generation. The potential productivity of these variants under intensive cultivation conditions was not proved [3]. Dwimahyani and Ishak [4] used induced mutations in J. curcas for improvement of agronomic characters with an irradiation dose of 10 Gy and identified mutant plants with early maturity, 100 seeds weight (30% over control) and better branch growth.
Different molecular techniques such as Amplified Fragment Length Polymorphism (AFLP) [5], Restriction Fragment Length Polymorphism (RFLP) [6], Simple Sequence Repeat (SSR) [7] and Randomly Amplified Polymorphic DNA (RAPD) [8] have been developed and are widely used in many fields of agriculture, medicine, forestry and forensic sciences etc., for various purposes. Among these, RAPD – an inexpensive and rapid method not requiring any information regarding the genome of the plant – has been widely used to document the genetic diversity in J. curcas [9–11] and other plants [12,13]. In India, mutation breeding using chemical and physical mutagens has been initiated to create genetic variation for various traits and developed mutants are being characterized using DNA markers [14]. Although the most common use of the RAPD marker analysis is related to genetic mapping, taxonomic and phylogenetic studies, the method has also been used to detect DNA alterations [15]. RAPD method has been used earlier to study genetic variability by radiation treatments in Chrysanthemum [16], sugarcane [17], groundnut [18], sunflower [19], cypress [20] and soybean [21]. The present study was therefore done to evaluate the effect of different doses of gamma irradiation on seed germination, flowering and fruit and yield traits of J. curcas and to identify DNA polymorphism among the mutants with PCR-RAPD marker analysis.
2 Materials and methods
2.1 Plant source, irradiation and morphology study
The seeds of J. curcas were treated with different doses of gamma rays viz., 0, 5, 10, 15, 20 and 25 Kr. The treated seeds were sown at 1 cm depth in plastic trays (23 × 27 cm, 6 cm in height) filled with river sand, red soil and farm yard manure in the ratio of 3:2:1. After a month of germination, twenty-five plants from each treatment were transferred to polythene bags containing 4 kg mixture of river sand, red soil and farm yard manure in the ratio of 3:2:1 and maintained up to 3 months. Thereafter, ten plants from each treatment were transplanted to experimental field. Observations were made for flowering (days to first flowering, days taken for flowering to fruiting, days taken for fruiting to maturity and total number of flower per inflorescence), fruit traits (number of fruits per bunch and number of fruit bunches per plant) and yield traits (days to first harvest, 100 seed weight and seed weight per plant). Data were analyzed using one way ANOVA in SPPS software package (17.0).
2.2 RAPD analysis
2.2.1 DNA isolation
The fresh leaf material was harvested from the ten-month-old plants treated with gamma rays. Genomic DNA was extracted by adopting the CTAB method outlined by Doyle and Doyle [22] with minor modifications. About 0.1 g of leaf tissue was submerged in absolute alcohol [23,24] for an hour instead of grinding it in liquid nitrogen and put in a 2 ml Eppendorf tube. To grind the sample 0.5 ml of extraction buffer (2% cetyl trimethyl ammonium bromide (CTAB), 100 mM Tris-HCl, 1.4 M NaCl, 20 mM ethylene diamine tetra acetic acid (EDTA) di-sodium salt, 0.2 M mercaptoethanol, 1% PVP, pH 8.0) was added and incubated at 65 °C for 60 min. The above sample was extracted with equal volume of chloroform: isoamylalcohol (24:1) and the supernatant was collected in a new tube and treated with RNase (10 mg/ml) for 30 min. at 37 °C. After incubation period, 3 M sodium acetate and absolute alcohol were added to the mixture and it was again incubated at −20 °C for an hour. The mixture was centrifuged at 3000 rpm for 5 min. The pellet was air-dried and dissolved in 100 μl of Milli Q water. The genomic DNA isolated was quantified spectrometrically by measuring absorbance at 260 nm and DNA was diluted to make a working solution for RAPD marker analysis.
2.2.2 PCR amplification
PCR amplification was performed twice for each primer to ensure their reproducibility in a total volume of 25 μl containing 20 mM Tris/HCl (pH 8.4), 50 mM KCl, 200 μM of each dNTP's (Sigma Aldrich, Bangalore, India), 2 mM MgCl2, 0.8 μM primer (Sigma Aldrich, Bangalore, India), 100 ng of template DNA and 0.5 U Taq DNA polymerase (Sigma Aldrich, Bangalore, India,) in Eppendorf Master Cycler personal (AG 22331 Hamburg, Germany) which was programmed to include predenaturation at 94 °C for 1 min., followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 35 °C for 1 min and extension at 72 °C for 1 min. The final cycle allowed an additional 5 min. period of extension at 72 °C. The PCR products were analyzed by electrophoresis through 2% agarose gels in 1X TBE buffer. After electrophoresis, the gel was stained in ethidium bromide and then visualized and the images were photographed using Biorad Gel Documentation System.
2.2.3 Data scoring and statistical analysis
Data of RAPD marker analysis were scored as discrete variables, using ‘1’ to indicate the presence and ‘0’ to indicate absence of bands for each primer. The faint and unclear bands were not considered for data scoring. The binary data so generated were used to estimate levels of polymorphism by dividing the polymorphic bands by the total number of scored bands. A dendrogram based on Jaccard similarity coefficients was constructed by using Unweighted Pair Group Method of Arithmetic means (UPGMA) with the SHAN module of NTSYS-PC 2.0 to show phenetic representation of genetic relationships as revealed by the similarity coefficient [25].
3 Results
3.1 Seed germination, flowering, fruit and yield traits in gamma rays treated plants
Table 1 shows that seed germination in the control as well as in the treated seeds of J. curcas started simultaneously five days after sowing (DAS). The difference in the values of seed germination percentage, due to radiation doses was highly significant. In case of seed germination percentage, the data shows that 10 Kr dose (80.66) has a stimulatory effect while 15 Kr dose recorded a reduced germination of 37% as compared to control and other treatments.
Effects of gamma radiation on seed germination percentage, flowering, fruit and yield traits of J. curcas at maturity.
| Dose | Seed germination (%) | Days to first flowering | Days taken for flowering to fruiting | Days taken for fruiting to maturity | Total number of flower/inflorescence | No. of fruit bunches/plant | No. of fruits/bunch | Days to first harvest | 100 seed weight (g) | Seed weight (g plant −1) |
| Control | 58.32 ± 6.42 | 306 ± 8.02 | 8 ± 1.00 | 56 ± 4.72 | 218 ± 23 | 11 ± 0.85 | 9.75 ± 1.25 | 354 ± 18.33 | 71 ± 6.65 | 186 ± 6.50 |
| 5 Kr | 54.66 ± 5.03 | 255 ± 21.54 | 6 ± 1.00 | 49 ± 3.5 | 260 ± 32.95 | 16 ± 1.63 | 14 ± 3.10 | 310 ± 21.73 | 92 ± 2.17 | 227 ± 19.65 |
| 10 Kr | 80.65 ± 5.50 | 281 ± 15.82 | 7 ± 1.52 | 57 ± 2.52 | 239 ± 20.43 | 13 ± 1.25 | 11.50 ± 2.38 | 346 ± 16.92 | 82 ± 7.54 | 204 ± 5.03 |
| 15 Kr | 37.00 ± 4.35 | 299 ± 10.06 | 9 ± 1.00 | 59 ± 3.05 | 201 ± 18.34 | 12 ± 2.05 | 12.97 ± 2.97 | 366 ± 6.42 | 64 ± 11.24 | 201 ± 7.93 |
| 20 Kr | 44.99 ± 2.00 | 294 ± 32.34 | 8 ± 1.00 | 59 ± 6.80 | 177 ± 33.81 | 10 ± 1.70 | 10.55 ± 1.21 | 362 ± 37.80 | 61 ± 4.36 | 186 ± 12.42 |
| 25 Kr | 56.99 ± 3.60 | 279 ± 27.79 | 9 ± 0.50 | 58 ± 1.52 | 141 ± 16.62 | 7 ± 1.25 | 6.75 ± 1.70 | 347 ± 25.99 | 59 ± 4.00 | 157 ± 2.51 |
Days to first flowering ranged from 255 to 306 days. The earliest flowering was observed when J. curcas was irradiated with 5 and 25 Kr doses of gamma radiation. A maximum decrease in days to first flowering (255 days) was recorded in 5 Kr followed by 25 Kr dose (279 days). All treatment was set flowers earlier than that of control, which took longer duration of 306 days for flowering. A maximum of 9 days and a minimum of 6 days were seen in all treatments and control to reach fruiting after initiation of flowering. Least time taken for flowering to fruiting was 6 days recorded in 5 Kr dose, while longer time was of 9 days taken by 15 and 25 Kr doses of gamma rays treated plants. Total number of flowers per inflorescence had a high degree of variability as 5 Kr dose had 260 flowers, which were almost two times compared to the 25 Kr dose of gamma irradiation which had only 141 flowers. A similar trend was also observed for days taken from fruiting to maturity. The days taken for fruiting to maturity were ranged from 49 to 59 days. The least time taken was 49 days by 5 Kr dose and the most time of 59 days was taken by 15 and 20 Kr dose gamma rays treated plants (Table 1). Number of fruit bunches per plant was recorded maximum in 5 Kr dose (16). A minimum number of fruit bunches per plant were observed in 25 Kr dose (7). The number of fruits per bunch of all treated and control plant were varied significantly. A maximum number of fruits per bunch were 14 recorded in 5 Kr dose, where as minimum number of fruits of 6.75 was seen in 25 Kr dose when compared to that of control and other treatments of gamma irradiation.
Hundred seed weight (g) and seed yield per plant (g) was ranged from 59–92 g and 194–227 g, respectively. The highest yield of 100 seed weight (92 g) and seed yield per plant (227 g) were obtained through the plants treated with 5 Kr dose and the lowest yield of 100 seed weight (59 g) and seed yield per plant (194 g) was recorded at 25 Kr gamma rays treated plants (Table 1). Analysis of variance for all traits studied except days to first flowering, days taken for flowering to fruiting and days taken for fruiting to maturity revealed that there was significant variation among the control and treatments at P > 0.05% level.
3.2 RAPD marker analysis in gamma rays treated plants
In our research, differences among the mutants improved by applying radiation to J. curcas were examined using RAPD marker from the view of polymerizing as comparison. Twenty-three primers were selected for amplification and data were analyzed for identifying DNA polymorphism showing in the gamma rays treated plants.
The control plant shows amplification at all the primers according to RAPD results. Five and 25 Kr mutants showing the improved agronomic traits viz., flowering, fruits and seed traits showed differences as amplification at 19 primers and 10, 15, and 20 Kr mutants showing differences for such traits were amplified in 15 primers only.
A total of 90 bands were scored of which 53 were polymorphic. Polymorphism of amplified fragments is caused by: (i) base substitutions or deletion in the priming sites, (ii) insertions that render priming sites too distant to support amplification or (iii) insertions or deletions that change the size of the amplified fragment [26]. The number of bands generated per primer varied from 1 to 8 and a minimum of 1 band was generated by the primers OPAW16, OPAK07, OPAK15, OPS01, OPAK20 and OPAL09, while the maximum of 8 bands were scored with OPU13, OPAB-15, OPF01 and OPAB11 followed by OPT18 and OPAW12 which produced 6 bands (Table 2). The percentage of polymorphism was ranged from 0.00 to 100%. Primers OPAK14 and OPAM06 showed 100% polymorphism while no polymorphic bands were observed in primers OPAW16, OPAK07, OPAL15, OPS01, OPAK20 and OPAL09, all of them produced only monomorphic bands.
List of primers, number of amplified products, polymorphic bands, monomorphic bands and polymorphism percentage.
| S. No | Primers | Sequence (5’–3’) | Total amplified products | Polymorphic bands | Monomorphic bands | Percent polymorphism |
| 1 | OPH18 | GAATCGGCCA | 2 | 1 | 1 | 50 |
| 2 | OPH12 | GGGACGTTGG | 5 | 3 | 2 | 60 |
| 3 | OPM13 | GGTGGTCAAG | 2 | 1 | 1 | 50 |
| 4 | OPM14 | AGGGTCGTTC | 3 | 2 | 1 | 66 |
| 5 | OPAL11 | GTCACGTCCT | 5 | 4 | 1 | 80 |
| 6 | OPT18 | CATGCCAGAC | 6 | 4 | 2 | 66 |
| 7 | OPA4 | AATCGGGCTG | 4 | 3 | 1 | 75 |
| 8 | OPAK14 | CTGTCATGCC | 2 | 2 | 0 | 100 |
| 9 | OPM15 | GACCTACCAC | 3 | 2 | 1 | 66 |
| 10 | OPU13 | GGCTGGTTCC | 8 | 5 | 3 | 62 |
| 11 | OPAB15 | CCTCCTTCTC | 8 | 2 | 6 | 25 |
| 12 | OPAW12 | GAGCAAGGCA | 6 | 2 | 4 | 33 |
| 13 | OPF01 | ACCGATCCTG | 8 | 2 | 6 | 25 |
| 14 | OPAW16 | TTACCCCGCT | 1 | – | 1 | – |
| 15 | OPAB11 | GTGCGCAATG | 8 | 7 | 1 | 87 |
| 16 | OPAK14 | CTGTCATGCC | 5 | 3 | 2 | 60 |
| 17 | OPF13 | GGCTGCAGAA | 5 | 4 | 1 | 80 |
| 18 | OPAK07 | CTTGGGGGAC | 1 | – | 1 | – |
| 19 | OPAK15 | ACCTGCCGTT | 1 | – | 1 | – |
| 20 | OPS01 | TCCTGGTCCC | 1 | – | 1 | – |
| 21 | OPAK20 | TGATGGCGTC | 1 | – | – | – |
| 22 | OPAM06 | TGGCGGTTTG | 4 | 4 | – | 100 |
| 23 | OPAL09 | TCTGCCAGTG | 1 | – | – | – |
| Total | 90 | 53 | 37 | 1285 | ||
| Mean | 3.9 | 2.3 | 1.6 | 55.86 |
From Table 3, it was seen that three other mutants, namely 10, 15 and 20 Kr were not more different from each other when compared to that of the control. Dendrogram (Fig. 1) indicated three distinct clusters, one comprising 4 treatments viz., control, 10, 15 and 20 Kr mutants while second and third cluster, included only one treatment each viz., 25 and 5 Kr mutants respectively which indicating its higher genetic distinctness from other treatments of gamma radiation. According to the resulting dendrogram, the 5 and 25 Kr mutants were more distant to the control than to the other mutants.
Distance matrix based on Jaccard coefficients.
| Control | 5 Kr | 10 Kr | 15 Kr | 20 Kr | 25 Kr | |
| Control | 0.000 | |||||
| 5 Kr | 0.397 | 0.000 | ||||
| 10 Kr | 0.297 | 0.426 | 0.000 | |||
| 15 Kr | 0.292 | 0.393 | 0.233 | 0.000 | ||
| 20 Kr | 0.286 | 0.390 | 0.283 | 0.250 | 0.000 | |
| 25 Kr | 0.371 | 0.456 | 0.345 | 0.339 | 0.632 | 0.000 |
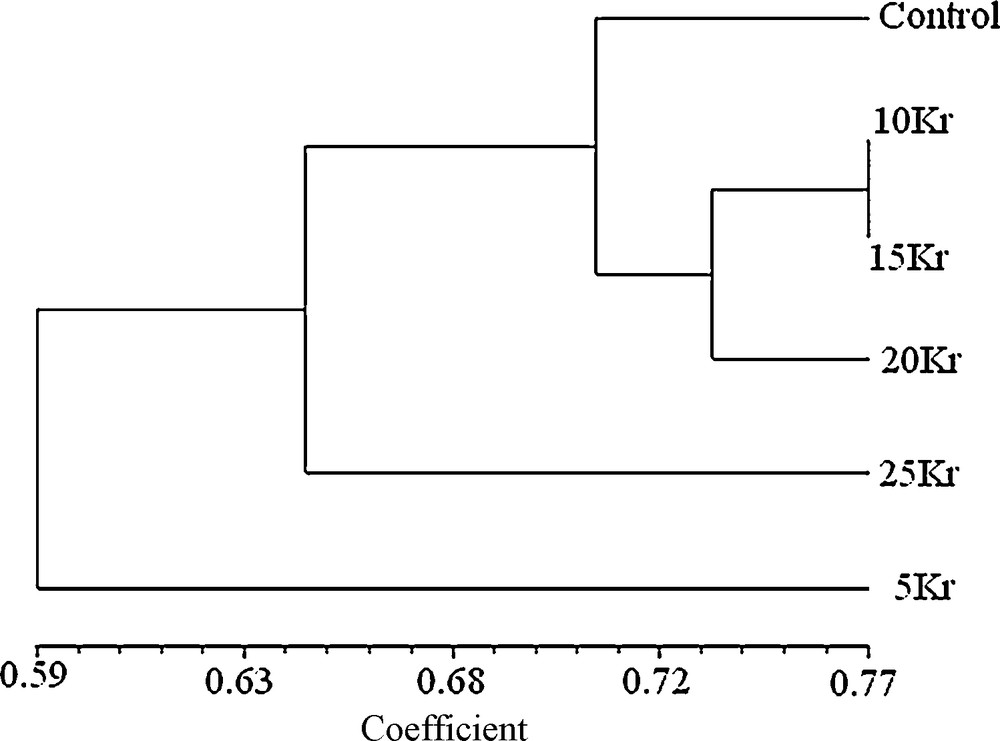
Dendogram constructed from similarity coefficients, showing the clustering of six mutants (Control, 5, 10, 15, 20 and 25 Kr).
4 Discussion
4.1 Germination, flowering, fruit and yield traits in the gamma rays treated plants
Percentage reduction/stimulation in seed germination might have been due to the effect of mutagens on meristematic tissues of the seed. The decrease in germination at higher doses of the mutagens may be attributed to disturbances at cellular level (caused either at physiological or physical level) including chromosomal damages. Kumar and Mishra [27] reported that in okra (Abelmoschus esculentus) germination percentage generally decreased with increasing gamma ray dose. Reduced germination percentage with increasing doses of gamma radiation was reported in Pinus [28], Rye [29], Chickpea [30,31] and Cowpea [32]. The higher exposures are usually inhibitor on seed germination of Gymnosperm and Angiosperm [33–35], where lower exposures are induced seed germination percentage [36,37]. The hypothetic origins of these stimulations are acceleration in cell division rates [38] as well as an activation of auxin [39]. The variation observed in reproductive characters such as days to first flowering, days taken for flowering to fruiting, days taken for fruiting to maturity and total number of flowers per inflorescence can be useful for plant cultivation with high yield as the primary objective. The findings on days to first flowering, days taken for flowering to fruiting, days taken for fruiting to maturity and total number of flowers per inflorescence showed that gamma rays treated plants can change the flowering and its maturity in either a positive or negative direction which result in sufficient variability in the treated population that can be utilized for selection of early or late flowering plants for further crop improvement. Days to flowering and maturity did not significantly vary with control and treated plants due to gamma rays. These two traits are very important in cultivation of J. curcas as it is a sensitive plant to flooding and water lodging conditions in winter season. Days to flowering and maturity in the case of irradiated populations (Table 1) were consistently shifted towards earliness. It is valuable in obtaining varieties associated with escape from pests, drought and other stress injuries that occur in late growing season. Difference in effect of gamma rays might be due to seed metabolism and onset of DNA synthesis [40]. The present results provide support to hypothesis of Gaul and Asistveit [41] and Larik et al. [42] which state that changes in mean values of quantitative characters occurs in the direction associated with reduced vitality independent of genotype used.
Considering the effect of physical mutagen on 100 seed weight and seed weight per plant, it is apparent that highly positive results obtained in lower dose of gamma radiation are suggestive of the usefulness of mutation breeding technique in J. curcas. The 5 Kr dose showed highest 100 seed weight and seed yield per plant. The 5 Kr dose exhibited maximum 100 seed weight and seed yield per plant than the control and other treatments, which probably indicate an increase in the size of seed as a result of induced mutation. This study shows that 5 Kr dose was a stimulative dose for flowering, fruits and seed yield in J. curcas. In several investigations report, such stimulation provoked with low doses of ionizing radiations in Pinus kesiya and P. wallichiana [28], Anacardium occidentale [43], Vitis vinifera [44], Eruca veisicaria [45], Pisum sativum [38] and Triticum durum [46]. The higher doses of gamma irradiation lead to an overall reduction in all parameters studied. This may be partly due to the fact that the cells which have relatively more chromosomal damage at high irradiation exposures are at a disadvantage due to diplontic section, which cannot complete well with the normal cells and are thus prevented from making any further contribution. Mutation induction technology has been used as a practical tool to develop new varieties through improving character of direct importance i.e., early maturity, tolerance to biotic and abiotic stresses. Gamma rays are used to obtain high frequency gene mutation and chromosomal alteration in different crops. According to the FAO database, a number of mutant varieties have been evolved through induced mutation.
4.2 RAPD marker analysis in the gamma rays treated plants
The aim of the present study was to evaluate DNA polymorphism through RAPD markers in J. curcas showing differences in agronomic traits studied by gamma radiation. The induction of mutations by ionizing radiation starts with the interaction of a radiation field with plants. The genetic alternations produced by ionizing radiation are due to ionization and excitations of the DNA molecule. A number of different types of chemical changes are induced. There are two effects of using radiation on the heredity material; gene mutations and chromosome breaks. Induced mutations contribute genetic variability by increasing DNA polymorphism and they are used in plant breeding.
Although the most common use of the RAPD marker analysis is related to genetic mapping, taxonomic and phylogenetic studies [8], the method has also been used to detect DNA alternation and mutation. The polymorphism in genomic DNA was detected by RAPD profiles through the randomly primed PCR reactions. In this sense, the obvious disappearance of normal bands and appearance of new bands generated from the plants exposed to different doses of gamma irradiation in comparison to the untreated control. In our study, the number of lost bands was found higher than that of extra bands. The lost bands in J. curcas plants exposed to gamma irradiation were determined generally as PCR products with different molecular weight. It is suggested that the DNA damage may be serious in the majority of cells in the plant parts of J. curcas exposed to gamma irradiation. The disappearance of normal bands (loss of bands) may be related to the event such as DNA damage (e.g. single and double-strand breaks, modified bases, abasic sits, oxidized bases, bulky adducts), DNA-protein cross links, point mutation and/or complex chromosomal rearrangements induced by gamma radiation. Radiation is one of the best known physical mutagen, as it dissociates the atoms of water molecules and causes the generation of hydroxyl radicals and oxidative damage [47] that are the most reactive. They react with most biomolecules including DNA and scavenge electrons from them. The oxidation of biomolecules by the radicles damage their structure and biological activity. Thus, genetic alterations occur on the DNA molecules. When Taq DNA polymerase encounters a DNA adduct, there are a number of possible outcomes including blockage, bypass and the possible dissociation of the enzyme/adduct complex, which will cause the loss of bands [48]. Appearance of new PCR products (extra bands) was also detected in RAPD profiles. New PCR amplification products may reveal a change in some oligonucleotide priming sites due to mutations [new annealing event(s)], large deletions (bringing to pre-existing annealing site closer), and/or homologous recombinations (juxtaposing two sequences that match the sequences of primer) [49]. Atienzar et al. [50] reported that mutations can only be responsible for the appearance of new bands if they occur at same locus in a sufficient number of cells (a minimum of 10% of mutations may be required to new PCR product visible in agarose gel) to be amplified by PCR. The new bands could be attributed to mutation, while the disappearance of bands could be attributed to DNA damage [48], both of them resulting in generation of DNA polymorphism in the gamma rays treated plants.
By physical mutagen, new varieties were improved in many plants with X-rays and gamma radiation. In these varieties, some important agronomical traits were improved such as flower and seed colour, early flowering, resistant to disease, salinity resistant to cold and heat resistant to lodging and yield quality, etc. [51]. The present study coincides with them by having obtained plants with some improved agronomical traits of early germination (10 Kr mutant), early flowering (5 and 25 Kr mutants), fruit and yield traits (5 Kr mutant) in J. curcas. Atak et al. [21] found that there was potential polymorphism and improvement between mutants and control while they determined the differences among mutants applying to various doses of atrazine and radiation in soybean varieties with RAPD marker. RAPD markers, which can quickly detect a large number of genetic polymorphism, have led to creation of genetic maps in a number of woody fruit crops [52] and sunflower [19] including changes due to DNA damage and RAPD markers have been used to detect mutations and DNA damage [53].
With the phenetic numerical analysis, the dendrogram constructed based on UPGMA analysis of RAPD data is one of the most effective methods in numerical computation and it can show the relationships of every sample [54]. In this sense, the phenetic approach differs from the genetic approach in that RAPD profiles are not considered as genotypes. In the present research, when markers obtained the end of RAPD analyses done at the plants showing improved agronomic traits and control were examined, it was seen that there was a potential polymorphism between mutants and controls. In the dendrograms done according to RAPD markers, the genetically distance to control and mutants replying to gamma radiation in various doses in particular, 5 Kr and 25 Kr were rather different.
5 Conclusion
For the purpose of plant breeding programme, mutagenic treatments with low physiological effects and strong genetic effects are desirable. Hence, we used gamma irradiation dose range of 5–25 Kr for crop improvement in J. curcas of which lower doses, particularly 5 Kr dose, was effective to influence the agronomical traits except seed germination percentage which was influenced by 10 Kr dose while other treatment such as 15, 20, and 25 Kr doses were to reduce the traits studied. This is the first report on molecular characterization of induced mutagenesis through gamma irradiation using RAPD marker analysis in J. curcas. Our data support the view that the RAPD analysis is a highly suitable method for the detection of DNA polymorphism induced by gamma radiation. The RAPD marker generated genetic variation among control and gamma rays treated populations will help to distinguish the plants showing differences in morphological characters.
Conflict of interest statement
There is no conflict of interest because of this is the first report in Jatropha curcas using gamma rays and RAPD marker.
Acknowledgements
The authors thank the University Grant Commission (UGC), New Delhi, India for providing the fund for the present study. We thank the authorities of Annamalai University and Dr. R. Panneerselvam, Professor and Head, Department of Botany, Annamalai University, for having provided laboratory facilities and encouragements. We also acknowledge Dr. N. Nalini, Professor, Department of Biochemistry and Biotechnology, Annamalai University, for rendering all help for using Gel Documentation.


